TCR-Engineered T Cells Directed against Ropporin-1 Constitute a Safe and Effective Treatment for Triple-Negative Breast Cancer
- PMID: 39172012
- PMCID: PMC11609630
- DOI: 10.1158/2159-8290.CD-24-0168
TCR-Engineered T Cells Directed against Ropporin-1 Constitute a Safe and Effective Treatment for Triple-Negative Breast Cancer
Abstract
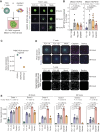
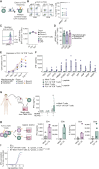
Triple-negative breast cancer (TNBC) has an urgent need for new therapies. We discovered Ropporin-1 (ROPN1) as a target to treat TNBC with T cells. ROPN1 showed high and homogenous expression in 90% of primary and metastatic TNBC but not in healthy tissues. Human leukocyte antigen-A2-binding peptides were detected via immunopeptidomics and predictions and used to retrieve T-cell receptors (TCR) from naïve repertoires. Following gene introduction into T cells and stringent selection, we retrieved a highly specific TCR directed against the epitope FLYTYIAKV that did not recognize noncognate epitopes from alternative source proteins. Notably, this TCR-mediated killing of three-dimensional (3D) tumoroids in vitro and tumor cells in vivo and outperformed standard-of-care drugs. Finally, the T-cell product expressing this TCR and manufactured using a clinical protocol fulfilled standard safety and efficacy assays. Collectively, we have identified and preclinically validated ROPN1 as a target and anti-ROPN1 TCR T cells as a treatment for the vast majority of patients with TNBC. Significance: Metastatic TNBC has a dismal prognosis. This study discovers Ropporin-1 as a target for T-cell therapy for most patients. The selected TCR is highly specific and sensitive in advanced models, and preclinical testing shows that the T-cell product expressing this TCR, manufactured according to good manufacturing practice, has favorable safety and potency.
©2024 The Authors; Published by the American Association for Cancer Research.
Conflict of interest statement
D. Hammerl reports grants from Dutch Cancer Society and Health Holland Public-private partnership during the conduct of the study and has a patent for P128827EP00 pending. M.T.A. de Beijer reports personal fees from the Dutch Ministry of Health, Welfare and Sport and personal fees from Erasmus University Medical Center outside the submitted work and has a patent for WO 2021/110919 pending to ISA Pharmaceuticals B.V. and Erasmus University Medical Center. S.I. Buschow reports grants and nonfinancial support from ISA Pharmaceuticals BV, and Merus BV, Pfizer Inc. and grants and nonfinancial support from Numab therapeutics AG outside the submitted work. M. Kok reports grants from Bristol Myers Squibb, Roche, and AZ, other support from MSD, AZ, and Gilead, and other support from BioNTech outside the submitted work. E.H.J. Danen reports grants from Genmab B.V. outside the submitted work. J.W.M. Martens reports grants from the Dutch Cancer Society during the conduct of the study and grants from the Dutch Cancer Society, Netherlands Organisation for Scientific Research (NWO), European Research Council, S. Komen Foundation, Erasmus University Medical Center Rotterdam, the Netherlands, breast cancer Now, MLDS NL, GSK, Pfizer, Philips, Cergentis, Therawis, Pamgene, Cytrotrack, and Menarini and personal fees from Novartis outside the submitted work. R.J.M. Abbott reports grants from Health∼Holland, other support from Swanbridge Capital, Van Herk Ventures, and Thuja Capital during the conduct of the study, and personal fees and other support from Enara Bio Ltd. and Adaptimmune Ltd. outside the submitted work. R. Debets reports grants from Health∼Holland Public-Private Partnership award EMC-TKI LSH20020, Erasmus MC Daniel den Hoed Foundation, and the Dutch Cancer Society KWF 2014-7087 during the conduct of the study and grants from MSD, Bayer, and Pan Cancer T outside the submitted work, as well as has patents for P128827EP00 and P136875EP00 pending and licensed to Pan Cancer T and a patent for P130556EP00 pending. No disclosures were reported by the other authors.
Figures




References
-
- Robson M, Im S-A, Senkus E, Xu B, Domchek SM, Masuda N, et al. . Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med 2017;377:523–33. - PubMed
-
- Kuchenbaecker KB, Hopper JL, Barnes DR, Phillips K-A, Mooij TM, Roos-Blom M-J, et al. . Risks of breast, ovarian, and contralateral breast cancer for BRCA1 and BRCA2 mutation carriers. JAMA 2017;317:2402–16. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

