Hypoxia-induced complement component 3 promotes aggressive tumor growth in the glioblastoma microenvironment
- PMID: 39172519
- PMCID: PMC11466187
- DOI: 10.1172/jci.insight.179854
Hypoxia-induced complement component 3 promotes aggressive tumor growth in the glioblastoma microenvironment
Abstract
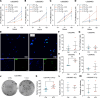
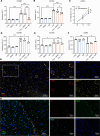
Glioblastoma (GBM) is the most aggressive form of glioma with a high rate of relapse despite intensive treatment. Tumor recurrence is tightly linked to radio-resistance, which in turn is associated with hypoxia. Here, we discovered a strong link between hypoxia and local complement signaling using publicly available bulk, single-cell, and spatially resolved transcriptomic data from patients with GBM. Complement component 3 (C3) and the receptor C3AR1 were both associated with aggressive disease and shorter survival in human glioma. In a genetically engineered mouse model of GBM, we found C3 specifically in hypoxic tumor areas. In vitro, we found an oxygen level-dependent increase in C3 and C3AR1 expression in response to hypoxia in several GBM and stromal cell types. C3a induced M2 polarization of cultured microglia and macrophages in a C3aR-dependent fashion. Targeting C3aR using the antagonist SB290157 prolonged survival of glioma-bearing mice both alone and in combination with radiotherapy while reducing the number of M2-polarized macrophages. Our findings establish a strong link between hypoxia and complement pathways in GBM and support a role of hypoxia-induced C3a/C3aR signaling as a contributor to glioma aggressiveness by regulating macrophage polarization.
Keywords: Cancer; Complement; Hypoxia; Oncology.
Conflict of interest statement
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

