DOT1L-mediated RAP80 methylation promotes BRCA1 recruitment to elicit DNA repair
- PMID: 39172790
- PMCID: PMC11363320
- DOI: 10.1073/pnas.2320804121
DOT1L-mediated RAP80 methylation promotes BRCA1 recruitment to elicit DNA repair
Abstract
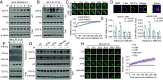
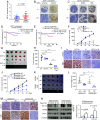
Breast Cancer Type 1 Susceptibility Protein (BRCA1) is a tumor-suppressor protein that regulates various cellular pathways, including those that are essential for preserving genome stability. One essential mechanism involves a BRCA1-A complex that is recruited to double-strand breaks (DSBs) by RAP80 before initiating DNA damage repair (DDR). How RAP80 itself is recruited to DNA damage sites, however, is unclear. Here, we demonstrate an intrinsic correlation between a methyltransferase DOT1L-mediated RAP80 methylation and BRCA1-A complex chromatin recruitment that occurs during cancer cell radiotherapy resistance. Mechanistically, DOT1L is quickly recruited onto chromatin and methylates RAP80 at multiple lysines in response to DNA damage. Methylated RAP80 is then indispensable for binding to ubiquitinated H2A and subsequently triggering BRCA1-A complex recruitment onto DSBs. Importantly, DOT1L-catalyzed RAP80 methylation and recruitment of BRCA1 have clinical relevance, as inhibition of DOT1L or RAP80 methylation seems to enhance the radiosensitivity of cancer cells both in vivo and in vitro. These data reveal a crucial role for DOT1L in DDR through initiating recruitment of RAP80 and BRCA1 onto chromatin and underscore a therapeutic strategy based on targeting DOT1L to overcome tumor radiotherapy resistance.
Keywords: BRCA1; DNA damage repair; DOT1L; RAP80; histone modification.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





References
-
- Lord C. J., Ashworth A., The DNA damage response and cancer therapy. Nature 481, 287–294 (2012). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

