Global transcriptome analysis identifies critical functional modules associated with multiple abiotic stress responses in microalgae Chromochloris zofingiensis
- PMID: 39172989
- PMCID: PMC11341014
- DOI: 10.1371/journal.pone.0307248
Global transcriptome analysis identifies critical functional modules associated with multiple abiotic stress responses in microalgae Chromochloris zofingiensis
Abstract
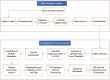
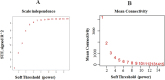
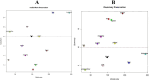
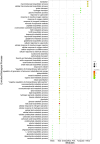
In the current study, systems biology approach was applied to get a deep insight regarding the regulatory mechanisms of Chromochloris zofingiensis under overall stress conditions. Meta-analysis was performed using p-values combination of differentially expressed genes. To identify the informative models related to stress conditions, two distinct weighted gene co-expression networks were constructed and preservation analyses were performed using medianRankand Zsummary algorithms. Moreover, functional enrichment analysis of non-preserved modules was performed to shed light on the biological performance of underlying genes in the non-preserved modules. In the next step, the gene regulatory networks between top hub genes of non-preserved modules and transcription factors were inferred using ensemble of trees algorithm. Results showed that the power of beta = 7 was the best soft-thresholding value to ensure a scale-free network, leading to the determination of 12 co-expression modules with an average size of 128 genes. Preservation analysis showed that the connectivity pattern of the six modules including the blue, black, yellow, pink, greenyellow, and turquoise changed during stress condition which defined as non-preserved modules. Examples of enriched pathways in non-preserved modules were Oxidative phosphorylation", "Vitamin B6 metabolism", and "Arachidonic acid metabolism". Constructed regulatory network between identified TFs and top hub genes of non-preserved module such as Cz06g10250, Cz03g12130 showed that some specific TFs such as C3H and SQUAMOSA promoter binding protein (SBP) specifically regulates the specific hubs. The current findings add substantially to our understanding of the stress responsive underlying mechanism of C. zofingiensis for future studies and metabolite production programs.
Copyright: © 2024 Bahman Panahi. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Razeghi J, Pishtab PA, Fathi P, Panahi B, Hejazi MA. The feasibility of microalgae Dunaliella identification based on conserved regions of mitochondrial cytochrome b and cytochrome oxidase genes. Cytology and Genetics. 2021;55(6):558–65.
-
- Panahi B, Farhadian M, Hejazi MA. Systems biology approach identifies functional modules and regulatory hubs related to secondary metabolites accumulation after transition from autotrophic to heterotrophic growth condition in microalgae. Plos one. 2020;15(2):e0225677. doi: 10.1371/journal.pone.0225677 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

