Members of an array of zinc-finger proteins specify distinct Hox chromatin boundaries
- PMID: 39173638
- PMCID: PMC11613955
- DOI: 10.1016/j.molcel.2024.08.007
Members of an array of zinc-finger proteins specify distinct Hox chromatin boundaries
Abstract
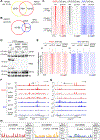
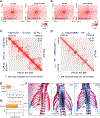
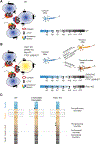
Partitioning of repressive from actively transcribed chromatin in mammalian cells fosters cell-type-specific gene expression patterns. While this partitioning is reconstructed during differentiation, the chromatin occupancy of the key insulator, CCCTC-binding factor (CTCF), is unchanged at the developmentally important Hox clusters. Thus, dynamic changes in chromatin boundaries must entail other activities. Given its requirement for chromatin loop formation, we examined cohesin-based chromatin occupancy without known insulators, CTCF and Myc-associated zinc-finger protein (MAZ), and identified a family of zinc-finger proteins (ZNFs), some of which exhibit tissue-specific expression. Two such ZNFs foster chromatin boundaries at the Hox clusters that are distinct from each other and from MAZ. PATZ1 was critical to the thoracolumbar boundary in differentiating motor neurons and mouse skeleton, while ZNF263 contributed to cervicothoracic boundaries. We propose that these insulating activities act with cohesin, alone or combinatorially, with or without CTCF, to implement precise positional identity and cell fate during development.
Keywords: CTCF; Hox genes in development; MAZ; PATZ1; chromatin domains; cohesin; gene regulation; genome organization; insulation; motor neurons; zinc-finger proteins.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests D.R. was a co-founder of Constellation Pharmaceuticals and Fulcrum Therapeutics. Currently, D.R. has no affiliation with either company.
Figures




Update of
-
Members of an array of zinc finger proteins specify distinct Hox chromatin boundaries.bioRxiv [Preprint]. 2024 Aug 4:2023.04.25.538167. doi: 10.1101/2023.04.25.538167. bioRxiv. 2024. Update in: Mol Cell. 2024 Sep 19;84(18):3406-3422.e6. doi: 10.1016/j.molcel.2024.08.007. PMID: 37162865 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

