Targeting the liver clock improves fibrosis by restoring TGF-β signaling
- PMID: 39173955
- PMCID: PMC7618256
- DOI: 10.1016/j.jhep.2024.07.034
Targeting the liver clock improves fibrosis by restoring TGF-β signaling
Abstract
Background & aims: Liver fibrosis is the major driver of hepatocellular carcinoma and liver disease-related death. Approved antifibrotic therapies are absent and compounds in development have limited efficacy. Increased TGF-β signaling drives collagen deposition by hepatic stellate cells (HSCs)/myofibroblasts. Here, we aimed to dissect the role of the circadian clock (CC) in controlling TGF-β signaling and liver fibrosis.
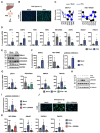
Methods: Using CC-mutant mice, enriched HSCs and myofibroblasts obtained from healthy and fibrotic mice in different CC phases and loss-of-function studies in human hepatocytes and myofibroblasts, we investigated the relationship between CC and TGF-β signaling. We explored hepatocyte-myofibroblast communication through bioinformatic analyses of single-nuclei transcriptomes and performed validation in cell-based models. Using mouse models for MASH (metabolic dysfunction-associated steatohepatitis)-related fibrosis and spheroids from patients with liver disease, we performed proof-of-concept studies to validate pharmacological targetability and clinical translatability.
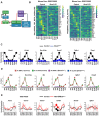
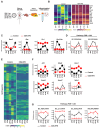
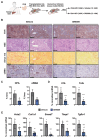
Results: We discovered that the CC oscillator temporally gates TGF-β signaling and this regulation is broken in fibrosis. We demonstrate that HSCs and myofibroblasts contain a functional CC with rhythmic expression of numerous genes, including fibrogenic genes. Perturbation studies in hepatocytes and myofibroblasts revealed a reciprocal relationship between TGF-β activation and CC perturbation, which was confirmed in patient-derived ex vivo and in vivo models. Pharmacological modulation of CC-TGF-β signaling inhibited fibrosis in mouse models in vivo as well as in patient-derived liver spheroids.
Conclusion: The CC regulates TGF-β signaling, and the breakdown of this control is associated with liver fibrosis in patients. Pharmacological proof-of-concept studies across different models have uncovered the CC as a novel therapeutic target for liver fibrosis - a growing unmet medical need.
Impact and implications: Liver fibrosis due to metabolic diseases is a global health challenge. Many liver functions are rhythmic throughout the day, being controlled by the circadian clock (CC). Here we demonstrate that regulation of the CC is perturbed upon chronic liver injury and this perturbation contributes to fibrotic disease. By showing that a compound targeting the CC improves liver fibrosis in patient-derived models, this study provides a novel therapeutic candidate strategy to treat fibrosis in patients. Additional studies will be needed for clinical translation. Since the findings uncover a previously undiscovered profibrotic mechanism and therapeutic target, the study is of interest for scientists investigating liver disease, clinical hepatologists and drug developers.
Keywords: Liver disease; MASLD; circadian clock; drug discovery; hepatic stellate cells; transcriptional regulation.
Copyright © 2024 European Association for the Study of the Liver. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Figures




References
-
- Hagström H, Nasr P, Ekstedt M, et al. Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol. 2017;67:1265–73. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

