Bounding the Softwired Parsimony Score of a Phylogenetic Network
- PMID: 39174812
- PMCID: PMC11341636
- DOI: 10.1007/s11538-024-01350-9
Bounding the Softwired Parsimony Score of a Phylogenetic Network
Abstract
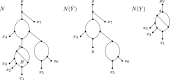
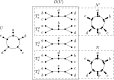
In comparison to phylogenetic trees, phylogenetic networks are more suitable to represent complex evolutionary histories of species whose past includes reticulation such as hybridisation or lateral gene transfer. However, the reconstruction of phylogenetic networks remains challenging and computationally expensive due to their intricate structural properties. For example, the small parsimony problem that is solvable in polynomial time for phylogenetic trees, becomes NP-hard on phylogenetic networks under softwired and parental parsimony, even for a single binary character and structurally constrained networks. To calculate the parsimony score of a phylogenetic network N, these two parsimony notions consider different exponential-size sets of phylogenetic trees that can be extracted from N and infer the minimum parsimony score over all trees in the set. In this paper, we ask: What is the maximum difference between the parsimony score of any phylogenetic tree that is contained in the set of considered trees and a phylogenetic tree whose parsimony score equates to the parsimony score of N? Given a gap-free sequence alignment of multi-state characters and a rooted binary level-k phylogenetic network, we use the novel concept of an informative blob to show that this difference is bounded by times the softwired parsimony score of N. In particular, the difference is independent of the alignment length and the number of character states. We show that an analogous bound can be obtained for the softwired parsimony score of semi-directed networks, while under parental parsimony on the other hand, such a bound does not hold.
Keywords: Level-k; Parental parsimony; Phylogenetic networks; Softwired parsimony.
© 2024. The Author(s).
Figures





References
-
- Cardona G, Rossello F, Valiente G (2009) Comparison of tree-child phylogenetic networks. IEEE/ACM Trans Comput Biol Bioinf 6(4):552–569 - PubMed
-
- Döcker J, Linz S, Semple C (2024) Hypercubes and Hamilton cycles of display sets of rooted phylogenetic networks. Adv Appl Math 152:102595
-
- Felsenstein J (2004) Inferring phylogenies. Sinauer Associates, New York
-
- Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20(4):406–416
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

