Targeted viromes and total metagenomes capture distinct components of bee gut phage communities
- PMID: 39175056
- PMCID: PMC11342477
- DOI: 10.1186/s40168-024-01875-0
Targeted viromes and total metagenomes capture distinct components of bee gut phage communities
Abstract
Background: Despite being among the most abundant biological entities on earth, bacteriophage (phage) remain an understudied component of host-associated systems. One limitation to studying host-associated phage is the lack of consensus on methods for sampling phage communities. Here, we compare paired total metagenomes and viral size fraction metagenomes (viromes) as methods for investigating the dsDNA viral communities associated with the GI tract of two bee species: the European honey bee Apis mellifera and the eastern bumble bee Bombus impatiens.
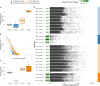
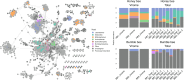
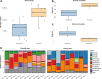
Results: We find that viromes successfully enriched for phage, thereby increasing phage recovery, but only in honey bees. In contrast, for bumble bees, total metagenomes recovered greater phage diversity. Across both bee species, viromes better sampled low occupancy phage, while total metagenomes were biased towards sampling temperate phage. Additionally, many of the phage captured by total metagenomes were absent altogether from viromes. Comparing between bees, we show that phage communities in commercially reared bumble bees are significantly reduced in diversity compared to honey bees, likely reflecting differences in bacterial titer and diversity. In a broader context, these results highlight the complementary nature of total metagenomes and targeted viromes, especially when applied to host-associated environments.
Conclusions: Overall, we suggest that studies interested in assessing total communities of host-associated phage should consider using both approaches. However, given the constraints of virome sampling, total metagenomes may serve to sample phage communities with the understanding that they will preferentially sample dominant and temperate phage. Video Abstract.
Keywords: Bacteriophage; Bee microbiome; Host-associated microbiome; Microbial ecology; Viral ecology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Honey bees harbor a diverse gut virome engaging in nested strain-level interactions with the microbiota.Proc Natl Acad Sci U S A. 2020 Mar 31;117(13):7355-7362. doi: 10.1073/pnas.2000228117. Epub 2020 Mar 16. Proc Natl Acad Sci U S A. 2020. PMID: 32179689 Free PMC article.
-
Global Composition of the Bacteriophage Community in Honey Bees.mSystems. 2022 Apr 26;7(2):e0119521. doi: 10.1128/msystems.01195-21. Epub 2022 Mar 28. mSystems. 2022. PMID: 35343797 Free PMC article.
-
Viromes vs. mixed community metagenomes: choice of method dictates interpretation of viral community ecology.bioRxiv [Preprint]. 2023 Dec 12:2023.10.15.562385. doi: 10.1101/2023.10.15.562385. bioRxiv. 2023. Update in: Microbiome. 2024 Oct 7;12(1):195. doi: 10.1186/s40168-024-01905-x. PMID: 37904928 Free PMC article. Updated. Preprint.
-
Functional and evolutionary insights into the simple yet specific gut microbiota of the honey bee from metagenomic analysis.Gut Microbes. 2013 Jan-Feb;4(1):60-5. doi: 10.4161/gmic.22517. Epub 2012 Oct 12. Gut Microbes. 2013. PMID: 23060052 Free PMC article. Review.
-
Does over a century of aerobic phage work provide a solid framework for the study of phages in the gut?Anaerobe. 2021 Apr;68:102319. doi: 10.1016/j.anaerobe.2021.102319. Epub 2021 Jan 16. Anaerobe. 2021. PMID: 33465423 Review.

