Genetic diversity and cross-species transmissibility of bat-associated picornaviruses from Spain
- PMID: 39175061
- PMCID: PMC11342490
- DOI: 10.1186/s12985-024-02456-1
Genetic diversity and cross-species transmissibility of bat-associated picornaviruses from Spain
Abstract
Background: Emerging zoonotic diseases arise from cross-species transmission events between wild or domesticated animals and humans, with bats being one of the major reservoirs of zoonotic viruses. Viral metagenomics has led to the discovery of many viruses, but efforts have mainly been focused on some areas of the world and on certain viral families.
Methods: We set out to describe full-length genomes of new picorna-like viruses by collecting feces from hundreds of bats captured in different regions of Spain. Viral sequences were obtained by high-throughput Illumina sequencing and analyzed phylogenetically to classify them in the context of known viruses. Linear discriminant analysis (LDA) was performed to infer likely hosts based on genome composition.
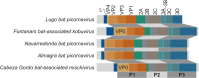
Results: We found five complete or nearly complete genomes belonging to the family Picornaviridae, including a new species of the subfamily Ensavirinae. LDA suggested that these were true vertebrate viruses, rather than viruses from the bat diet. Some of these viruses were related to picornaviruses previously found in other bat species from distant geographical regions. We also found a calhevirus genome that most likely belongs to a proposed new family within the order Picornavirales, and for which genome composition analysis suggested a plant host.
Conclusions: Our findings describe new picorna-like viral species and variants circulating in the Iberian Peninsula, illustrate the wide geographical distribution and interspecies transmissibility of picornaviruses, and suggest new hosts for calheviruses.
Keywords: Bat viruses; Calheviruses; Host inference; Interspecies transmission; Phylogenetics; Picornaviruses; Viral diversity; Viral metagenomics; Virus evolution.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Highly diverse population of Picornaviridae and other members of the Picornavirales, in Cameroonian fruit bats.BMC Genomics. 2017 Mar 23;18(1):249. doi: 10.1186/s12864-017-3632-7. BMC Genomics. 2017. PMID: 28335731 Free PMC article.
-
No Exchange of Picornaviruses in Vietnam between Humans and Animals in a High-Risk Cohort with Close Contact despite High Prevalence and Diversity.Viruses. 2021 Aug 27;13(9):1709. doi: 10.3390/v13091709. Viruses. 2021. PMID: 34578290 Free PMC article.
-
Full-genome sequencing of dozens of new DNA viruses found in Spanish bat feces.Microbiol Spectr. 2024 Aug 6;12(8):e0067524. doi: 10.1128/spectrum.00675-24. Epub 2024 Jul 11. Microbiol Spectr. 2024. PMID: 38990026 Free PMC article.
-
Avian picornaviruses: molecular evolution, genome diversity and unusual genome features of a rapidly expanding group of viruses in birds.Infect Genet Evol. 2014 Dec;28:151-66. doi: 10.1016/j.meegid.2014.09.027. Epub 2014 Sep 30. Infect Genet Evol. 2014. PMID: 25278047 Review.
-
Picornaviridae-the ever-growing virus family.Arch Virol. 2018 Feb;163(2):299-317. doi: 10.1007/s00705-017-3614-8. Epub 2017 Oct 20. Arch Virol. 2018. PMID: 29058149 Review.
Cited by
-
Identification and characterization of novel bat coronaviruses in Spain.PLoS Pathog. 2025 Aug 4;21(8):e1013371. doi: 10.1371/journal.ppat.1013371. eCollection 2025 Aug. PLoS Pathog. 2025. PMID: 40758759 Free PMC article.
-
Complete Genomes of DNA Viruses in Fecal Samples from Small Terrestrial Mammals in Spain.Viruses. 2024 Dec 5;16(12):1885. doi: 10.3390/v16121885. Viruses. 2024. PMID: 39772193 Free PMC article.
References
Publication types
MeSH terms
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

