Unveiling potential drug targets for lung squamous cell carcinoma through the integration of druggable genome and genome-wide association data
- PMID: 39175755
- PMCID: PMC11338847
- DOI: 10.3389/fgene.2024.1431684
Unveiling potential drug targets for lung squamous cell carcinoma through the integration of druggable genome and genome-wide association data
Abstract
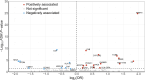
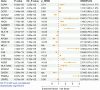
Background: Lung squamous cell carcinoma (LSCC) is a major subtype of lung cancer with poor prognosis and low survival rate. Compared with lung adenocarcinoma, yet no FDA-approved targeted-therapy has been found for lung squamous cell carcinoma. Methods: To identify potential drug targets for LSCC, Summary-data-based Mendelian randomization (SMR) analysis was used to examine the potential association between 4,543 druggable genes and LSCC, followed by colocalization analysis and HEIDI tests to confirm the robustness of the result. Phenome-wide association study (PheWAS) explored potential side effects of candidate drug targets. Enrichment analysis and protein-protein interaction networks revealed the function and significance of therapeutic targets. Single-cell expression analysis was used to examine cell types with enrichment expression of druggable genes in LSCC tissue. Drug prediction included screening potential drug candidates and evaluating their interactions with targets through molecular docking. Results: This research has identified ten significant drug targets for LSCC through a comprehensive SMR analysis. These targets included (COPA, PKD2L1, CCR1, C2, CYP21A2, and NCSTN as risk factors, and CCNA2, C4A, APOM, and LPAR2 as protective factors). PheWAS demonstrated that C2, CCNA2, LPAR2, and NCSTN exhibited associations with other phenotypes at the genetic level. Then, we found four potentially effective drugs with the Dsigdb database. Subsequently, molecular docking indicated that favorable binding interactions between drug candidates and potential target molecules. In the druggability evaluation, five out of ten drug target genes have been used in drug development (APOM, C4A, CCNA2, COPA, and PKD2L1). Six out of ten druggable genes showed significant expression in LSCC tissues (COPA, PKD2L1, CCR1, C2, NCSTN, LPAR2). Besides, Single-cell expression analysis revealed that C2 and CCNA2 were primarily enriched in macrophages, while COPA and NCSTN were enriched in both macrophages and epithelial cells. Conclusion: Our research revealed ten potential druggable genes for LSCC treatment, which might help to advance the precise and efficient therapeutic approaches of LSCC.
Keywords: drug targets; genome-wide association study; genomics; lung squamous cell carcinoma; summary-data-based mendelian randomization.
Copyright © 2024 Wu, Chen, Wen and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
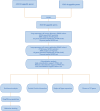
Figures





References
-
- Bao H., Li X., Cao Z., Huang Z., Chen L., Wang M., et al. (2022). Identification of COPA as a potential prognostic biomarker and pharmacological intervention target of cervical cancer by quantitative proteomics and experimental verification. J. Transl. Med. 20, 18. 10.1186/s12967-021-03218-1 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

