Metabolic Assessment in Non-Dialysis Patients with Chronic Kidney Disease
- PMID: 39176038
- PMCID: PMC11339343
- DOI: 10.2147/JIR.S461621
Metabolic Assessment in Non-Dialysis Patients with Chronic Kidney Disease
Abstract
Purpose: The aim of this study was to investigate the changes of different metabolites in the body fluids of non-dialysis patients with chronic kidney disease (CKD) using a metabolomics approach. The goal was to identify early biomarkers of CKD progression through metabolic pathway analysis.
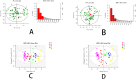
Patients and methods: Plasma samples from 47 patients with stages 1-4 CKD not requiring dialysis and 30 healthy controls were analyzed by liquid chromatography-mass spectrometry (LC-MS). Using multivariate data analysis, specifically a partially orthogonal least squares discriminant analysis model (OPLS-DA), we investigated metabolic differences between different stages of CKD. The sensitivity and specificity of the analysis were evaluated using the Area Under Curve (AUC) method. Furthermore, the metabolic pathways were analyzed using the Met PA database.
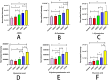
Results: Plasma samples from CKD patients and controls were successfully differentiated using an OPLS-DA model. Initially, twenty-five compounds were identified as potential plasma metabolic markers for distinguishing CKD patients from healthy controls. Among these, six compounds (ADMA, D-Ornithine, Kynurenine, Kynurenic acid, 5-Hydroxyindoleacetic acid, and Gluconic acid) were found to be associated with CKD progression It has been found to be associated with the progression of CKD. Changes in metabolic pathways associated with CKD progression include arginine and ornithine metabolism, tryptophan metabolism, and the pentose phosphate pathway.
Conclusion: By analyzing the metabolic pathways of different metabolites, we have identified the significant impact of CKD progression. The main metabolic pathways involved are Arginine and Ornithine metabolism, Tryptophan metabolism, and Pentose phosphate pathway. ADMA, D-Ornithine, L-Kynurenine, Kynurenic acid, 5-Hydroxyindoleacetic acid, and Gluconic acid could serve as potential early biomarkers for CKD progression. These findings have important implications for the early intervention and treatment of CKD, as well as for further research into the underlying mechanisms of its pathogenesis.
Keywords: arginine and ornithine metabolism; biomarkers; pentose phosphate pathway; tryptophan metabolism.
© 2024 Hong et al.
Conflict of interest statement
All authors report no conflicts of interest in this work.
Figures





References
LinkOut - more resources
Full Text Sources
Miscellaneous

