Genomic and Epigenomic Analysis of Plasma Cell-Free DNA Identifies Stemness Features Associated with Worse Survival in Lethal Prostate Cancer
- PMID: 39177583
- PMCID: PMC11743868
- DOI: 10.1158/1078-0432.CCR-24-1658
Genomic and Epigenomic Analysis of Plasma Cell-Free DNA Identifies Stemness Features Associated with Worse Survival in Lethal Prostate Cancer
Abstract
Purpose: Metastatic castration-resistant prostate cancer (mCRPC) resistant to androgen receptor signaling inhibitors (ARSI) is often lethal. Liquid biopsy biomarkers for this deadly form of disease remain under investigation, and underpinning mechanisms remain ill-understood.
Experimental design: We applied targeted cell-free DNA (cfDNA) sequencing to 126 patients with mCRPC from three academic cancer centers and separately performed genome-wide cfDNA methylation sequencing on 43 plasma samples collected prior to the initiation of first-line ARSI treatment. To analyze the genome-wide sequencing data, we performed nucleosome positioning and differential methylated region analysis. We additionally analyzed single-cell and bulk RNA sequencing data from 14 and 80 patients with mCRPC, respectively, to develop and validate a stem-like signature, which we inferred from cfDNA.
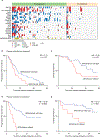
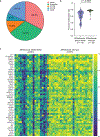
Results: Targeted cfDNA sequencing detected AR/enhancer alterations prior to first-line ARSIs that correlated with significantly worse progression-free survival (P = 0.01; HR = 2.12) and overall survival (P = 0.02; HR = 2.48). Plasma methylome analysis revealed that AR/enhancer lethal mCRPC patients have significantly higher promoter-level hypomethylation than AR/enhancer wild-type mCRPC patients (P < 0.0001). Moreover, gene ontology and CytoTRACE analysis of nucleosomally more accessible transcription factors in cfDNA revealed enrichment for stemness-associated transcription factors in patients with lethal mCRPC. The resulting stemness signature was then validated in a completely held-out cohort of 80 patients with mCRPC profiled by tumor RNA sequencing.
Conclusions: We analyzed a total of 220 patients with mCRPC, validated the importance of cell-free AR/enhancer alterations as a prognostic biomarker in lethal mCRPC, and showed that the underlying mechanism for lethality involves reprogramming developmental states toward increased stemness. See related commentary by Nawfal et al., p. 7.
©2024 American Association for Cancer Research.
Conflict of interest statement
Declaration of Interests
Figures



Update of
-
Genomic and epigenomic analysis of plasma cell-free DNA identifies stemness features associated with worse survival in AR -altered lethal prostate cancer.medRxiv [Preprint]. 2023 Dec 1:2023.12.01.23299215. doi: 10.1101/2023.12.01.23299215. medRxiv. 2023. Update in: Clin Cancer Res. 2025 Jan 6;31(1):151-163. doi: 10.1158/1078-0432.CCR-24-1658. PMID: 38077092 Free PMC article. Updated. Preprint.
Comment in
-
Advances in genomic biomarkers for lethal metastatic castration-resistant prostate cancer.Transl Cancer Res. 2025 Jun 30;14(6):3281-3285. doi: 10.21037/tcr-2025-232. Epub 2025 Jun 25. Transl Cancer Res. 2025. PMID: 40687240 Free PMC article. No abstract available.
References
-
- Fizazi K, Scher HI, Molina A, Logothetis CJ, Chi KN, Jones RJ, et al. Abiraterone acetate for treatment of metastatic castration-resistant prostate cancer: final overall survival analysis of the COU-AA-301 randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol 2012;13(10):983–92 doi 10.1016/S1470-2045(12)70379-0. - DOI - PubMed
-
- Ryan CJ, Smith MR, Fizazi K, Saad F, Mulders PF, Sternberg CN, et al. Abiraterone acetate plus prednisone versus placebo plus prednisone in chemotherapy-naive men with metastatic castration-resistant prostate cancer (COU-AA-302): final overall survival analysis of a randomised, double-blind, placebo-controlled phase 3 study. Lancet Oncol 2015;16(2):152–60 doi 10.1016/S1470-2045(14)71205-7. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

