Transforming Digital Phenotyping Raw Data Into Actionable Biomarkers, Quality Metrics, and Data Visualizations Using Cortex Software Package: Tutorial
- PMID: 39178032
- PMCID: PMC11380059
- DOI: 10.2196/58502
Transforming Digital Phenotyping Raw Data Into Actionable Biomarkers, Quality Metrics, and Data Visualizations Using Cortex Software Package: Tutorial
Abstract
As digital phenotyping, the capture of active and passive data from consumer devices such as smartphones, becomes more common, the need to properly process the data and derive replicable features from it has become paramount. Cortex is an open-source data processing pipeline for digital phenotyping data, optimized for use with the mindLAMP apps, which is used by nearly 100 research teams across the world. Cortex is designed to help teams (1) assess digital phenotyping data quality in real time, (2) derive replicable clinical features from the data, and (3) enable easy-to-share data visualizations. Cortex offers many options to work with digital phenotyping data, although some common approaches are likely of value to all teams using it. This paper highlights the reasoning, code, and example steps necessary to fully work with digital phenotyping data in a streamlined manner. Covering how to work with the data, assess its quality, derive features, and visualize findings, this paper is designed to offer the reader the knowledge and skills to apply toward analyzing any digital phenotyping data set. More specifically, the paper will teach the reader the ins and outs of the Cortex Python package. This includes background information on its interaction with the mindLAMP platform, some basic commands to learn what data can be pulled and how, and more advanced use of the package mixed with basic Python with the goal of creating a correlation matrix. After the tutorial, different use cases of Cortex are discussed, along with limitations. Toward highlighting clinical applications, this paper also provides 3 easy ways to implement examples of Cortex use in real-world settings. By understanding how to work with digital phenotyping data and providing ready-to-deploy code with Cortex, the paper aims to show how the new field of digital phenotyping can be both accessible to all and rigorous in methodology.
Keywords: Cortex; app; apps; clinical; data analysis; data processing; data set; data visualization; digital phenotyping; mental health; methodology; mindLAMP; mobile phone; open-source; real world; smartphone; smartphones.
©James Burns, Kelly Chen, Matthew Flathers, Danielle Currey, Natalia Macrynikola, Aditya Vaidyam, Carsten Langholm, Ian Barnett, Andrew (Jin Soo) Byun, Erlend Lane, John Torous. Originally published in the Journal of Medical Internet Research (https://www.jmir.org), 23.08.2024.
Conflict of interest statement
Conflicts of Interest: JT is the editor-in-chief of
Figures




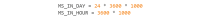

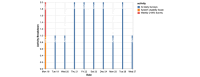






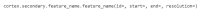
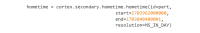
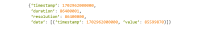
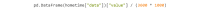
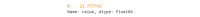
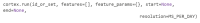
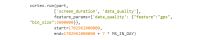

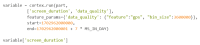








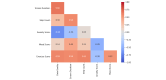
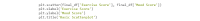
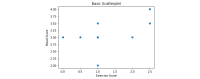
References
-
- Montag C, Quintana DS. Digital phenotyping in molecular psychiatry-a missed opportunity? Mol Psychiatry. 2023 Jan;28(1):6–9. doi: 10.1038/s41380-022-01795-1. https://europepmc.org/abstract/MED/36171355 10.1038/s41380-022-01795-1 - DOI - PMC - PubMed
-
- Moura I, Teles A, Viana D, Marques J, Coutinho L, Silva F. Digital phenotyping of mental health using multimodal sensing of multiple situations of interest: a systematic literature review. J Biomed Inform. 2023 Feb;138:104278. doi: 10.1016/j.jbi.2022.104278. https://linkinghub.elsevier.com/retrieve/pii/S1532-0464(22)00283-0 S1532-0464(22)00283-0 - DOI - PubMed
-
- Onnela JP. Opportunities and challenges in the collection and analysis of digital phenotyping data. Neuropsychopharmacology. 2021 Jan;46(1):45–54. doi: 10.1038/s41386-020-0771-3. https://europepmc.org/abstract/MED/32679583 10.1038/s41386-020-0771-3 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

