CD4+ and CD8+ T cells are required to prevent SARS-CoV-2 persistence in the nasal compartment
- PMID: 39178263
- PMCID: PMC11343035
- DOI: 10.1126/sciadv.adp2636
CD4+ and CD8+ T cells are required to prevent SARS-CoV-2 persistence in the nasal compartment
Abstract
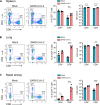
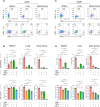
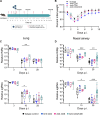
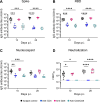
SARS-CoV-2 infection induces the generation of virus-specific CD4+ and CD8+ effector and memory T cells. However, the contribution of T cells in controlling SARS-CoV-2 during infection is not well understood. Following infection of C57BL/6 mice, SARS-CoV-2-specific CD4+ and CD8+ T cells are recruited to the respiratory tract, and a vast proportion secrete the cytotoxic molecule granzyme B. Using depleting antibodies, we found that T cells within the lungs play a minimal role in viral control, and viral clearance occurs in the absence of both CD4+ and CD8+ T cells through 28 days postinfection. In the nasal compartment, depletion of both CD4+ and CD8+ T cells, but not individually, results in persistent, culturable virus replicating in the nasal epithelial layer through 28 days postinfection. Viral sequencing analysis revealed adapted mutations across the SARS-CoV-2 genome, including a large deletion in ORF6. Overall, our findings highlight the importance of T cells in controlling virus replication within the respiratory tract during SARS-CoV-2 infection.
Figures




Update of
-
CD4+ and CD8+ T cells are required to prevent SARS-CoV-2 persistence in the nasal compartment.bioRxiv [Preprint]. 2024 Jan 24:2024.01.23.576505. doi: 10.1101/2024.01.23.576505. bioRxiv. 2024. Update in: Sci Adv. 2024 Aug 23;10(34):eadp2636. doi: 10.1126/sciadv.adp2636. PMID: 38410446 Free PMC article. Updated. Preprint.
References
-
- Ahn J. H., Kim J., Hong S. P., Choi S. Y., Yang M. J., Ju Y. S., Kim Y. T., Kim H. M., Rahman M. D. T., Chung M. K., Hong S. D., Bae H., Lee C. S., Koh G. Y., Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J. Clin. Invest. 131, e148517 (2021). - PMC - PubMed
-
- Lamers M. M., Haagmans B. L., SARS-CoV-2 pathogenesis. Nat. Rev. Microbiol. 20, 270–284 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

