Identification of a genetic region linked to tolerance to MRSA infection using Collaborative Cross mice
- PMID: 39178306
- PMCID: PMC11407622
- DOI: 10.1371/journal.pgen.1011378
Identification of a genetic region linked to tolerance to MRSA infection using Collaborative Cross mice
Abstract
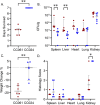
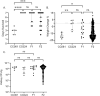
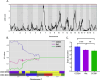
Staphylococcus aureus (S. aureus) colonizes humans asymptomatically but can also cause opportunistic infections, ranging from mild skin infections to severe life-threatening conditions. Resistance and tolerance are two ways a host can survive an infection. Resistance is limiting the pathogen burden, while tolerance is limiting the health impact of a given pathogen burden. In previous work, we established that collaborative cross (CC) mouse line CC061 is highly susceptible to Methicillin-resistant S. aureus infection (MRSA, USA300), while CC024 is tolerant. To identify host genes involved in tolerance after S. aureus infection, we crossed CC061 mice and CC024 mice to generate F1 and F2 populations. Survival after MRSA infection in the F1 and F2 generations was 65% and 55% and followed a complex dominant inheritance pattern for the CC024 increased survival phenotype. Colonization in F2 animals was more extreme than in their parents, suggesting successful segregation of genetic factors. We identified a Quantitative Trait Locus (QTL) peak on chromosome 7 for survival and weight change after infection. In this QTL, the WSB/EiJ (WSB) allele was present in CC024 mice and contributed to their MRSA tolerant phenotype. Two genes, C5ar1 and C5ar2, have high-impact variants in this region. C5ar1 and C5ar2 are receptors for the complement factor C5a, an anaphylatoxin that can trigger a massive immune response by binding to these receptors. We hypothesize that C5a may have altered binding to variant receptors in CC024 mice, reducing damage caused by the cytokine storm and resulting in the ability to tolerate a higher pathogen burden and longer survival.
Copyright: © 2024 Nagarajan et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



Similar articles
-
Collaborative Cross mice have diverse phenotypic responses to infection with Methicillin-resistant Staphylococcus aureus USA300.PLoS Genet. 2024 May 2;20(5):e1011229. doi: 10.1371/journal.pgen.1011229. eCollection 2024 May. PLoS Genet. 2024. PMID: 38696518 Free PMC article.
-
Community-acquired methicillin-resistant Staphylococcus aureus provoked cytokine storm causing severe infection on BALB/c mice.Mol Immunol. 2021 Dec;140:167-174. doi: 10.1016/j.molimm.2021.10.013. Epub 2021 Oct 28. Mol Immunol. 2021. PMID: 34717146
-
Extracellular vesicles from methicillin resistant Staphylococcus aureus stimulate proinflammatory cytokine production and trigger IgE-mediated hypersensitivity.Emerg Microbes Infect. 2021 Dec;10(1):2000-2009. doi: 10.1080/22221751.2021.1991239. Emerg Microbes Infect. 2021. PMID: 34623928 Free PMC article.
-
Methicillin-resistant Staphylococcus aureus: a controversial food-borne pathogen.Lett Appl Microbiol. 2017 Jun;64(6):409-418. doi: 10.1111/lam.12735. Epub 2017 May 3. Lett Appl Microbiol. 2017. PMID: 28304109 Review.
-
Into the storm: Chasing the opportunistic pathogen Staphylococcus aureus from skin colonisation to life-threatening infections.Environ Microbiol. 2017 Oct;19(10):3823-3833. doi: 10.1111/1462-2920.13833. Epub 2017 Jul 24. Environ Microbiol. 2017. PMID: 28631399 Review.
References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

