Fitness factor genes conserved within the multi-species core genome of Gram-negative Enterobacterales species contribute to bacteremia pathogenesis
- PMID: 39178317
- PMCID: PMC11376589
- DOI: 10.1371/journal.ppat.1012495
Fitness factor genes conserved within the multi-species core genome of Gram-negative Enterobacterales species contribute to bacteremia pathogenesis
Abstract
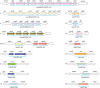
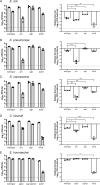
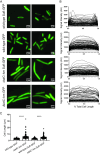
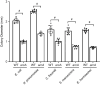
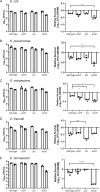
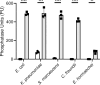
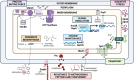
There is a critical gap in knowledge about how Gram-negative bacterial pathogens, using survival strategies developed for other niches, cause lethal bacteremia. Facultative anaerobic species of the Enterobacterales order are the most common cause of Gram-negative bacteremia, including Escherichia coli, Klebsiella pneumoniae, Serratia marcescens, Citrobacter freundii, and Enterobacter hormaechei. Bacteremia often leads to sepsis, a life-threatening organ dysfunction resulting from unregulated immune responses to infection. Despite a lack of specialization for this host environment, Gram-negative pathogens cause nearly half of bacteremia cases annually. Based on our existing Tn-Seq fitness factor data from a murine model of bacteremia combined with comparative genomics of the five Enterobacterales species above, we prioritized 18 conserved fitness genes or operons for further characterization. Mutants were constructed for all genes in all five species. Each mutant was used to cochallenge C57BL/6 mice via tail vein injection along with each respective wild-type strain to determine competitive indices for each fitness gene. Five fitness factor genes, when mutated, attenuated mutants in four or five species in the spleen and liver (tatC, ruvA, gmhB, wzxE, arcA). Five additional fitness factor genes or operons were validated as outcompeted by wild-type in three, four, or five bacterial species in the spleen (xerC, prc, apaGH, atpG, aroC). Overall, 17 of 18 fitness factor mutants were attenuated in at least one species in the spleen or liver. Together, these findings allow for the development of a model of bacteremia pathogenesis that may include future targets of therapy against bloodstream infections.
Copyright: © 2024 Mobley et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Commission TJ. Sepsis 2022. [Available from: https://www.jointcommission.org/resources/patient-safety-topics/infectio....
-
- Holmes CL, Albin OR, Mobley HLT, Bachman MA. Bloodstream Infections: Mechanisms of Pathogenesis and Opportunities for Intervention. Nature Reviews Microbiology. 2024;in press.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

