Combined Transcriptome and Circulating Tumor DNA Longitudinal Biomarker Analysis Associates With Clinical Outcomes in Advanced Solid Tumors Treated With Pembrolizumab
- PMID: 39178369
- PMCID: PMC11371115
- DOI: 10.1200/PO.24.00100
Combined Transcriptome and Circulating Tumor DNA Longitudinal Biomarker Analysis Associates With Clinical Outcomes in Advanced Solid Tumors Treated With Pembrolizumab
Abstract
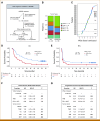
Purpose: Immune gene expression signatures are emerging as potential biomarkers for immunotherapy (IO). VIGex is a 12-gene expression classifier developed in both nCounter (Nanostring) and RNA sequencing (RNA-seq) assays and analytically validated across laboratories. VIGex classifies tumor samples into hot, intermediate-cold (I-Cold), and cold subgroups. VIGex-Hot has been associated with better IO treatment outcomes. Here, we investigated the performance of VIGex and other IO biomarkers in an independent data set of patients treated with pembrolizumab in the INSPIRE phase II clinical trial (ClinicalTrials.gov identifier: NCT02644369).
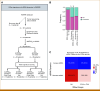
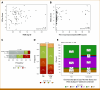
Materials and methods: Patients with advanced solid tumors were treated with pembrolizumab 200 mg IV once every 3 weeks. Tumor RNA-seq data from baseline tumor samples were classified by the VIGex algorithm. Circulating tumor DNA (ctDNA) was measured at baseline and start of cycle 3 using the bespoke Signatera assay. VIGex-Hot was compared with VIGex I-Cold + Cold and four groups were defined on the basis of the combination of VIGex subgroups and the change in ctDNA at cycle 3 from baseline (ΔctDNA).
Results: Seventy-six patients were enrolled, including 16 ovarian, 12 breast, 12 head and neck cancers, 10 melanoma, and 26 other tumor types. Objective response rate was 24% in VIGex-Hot and 10% in I-Cold/Cold. VIGex-Hot subgroup was associated with higher overall survival (OS) and progression-free survival (PFS) when included in a multivariable model adjusted for tumor type, tumor mutation burden, and PD-L1 immunohistochemistry. The addition of ΔctDNA improved the predictive performance of the baseline VIGex classification for both OS and PFS.
Conclusion: Our data indicate that the addition of ΔctDNA to baseline VIGex may refine prediction for IO.
Conflict of interest statement
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (
Figures


References
-
- André T, Shiu KK, Kim TW, et al. : Pembrolizumab in microsatellite-instability–high advanced colorectal cancer. N Engl J Med 383:2207-2218, 2020 - PubMed
-
- Marabelle A, Fakih M, Lopez J, et al. : Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 21:1353-1365, 2020 - PubMed
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical
Research Materials

