Altered memory CCR6+ Th17-polarised T-cell function and biology in people with HIV under successful antiretroviral therapy and HIV elite controllers
- PMID: 39178742
- PMCID: PMC11388266
- DOI: 10.1016/j.ebiom.2024.105274
Altered memory CCR6+ Th17-polarised T-cell function and biology in people with HIV under successful antiretroviral therapy and HIV elite controllers
Abstract
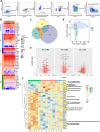
Background: Despite successful antiretroviral therapy (ART), frequencies and immunological functions of memory CCR6+ Th17-polarised CD4+ T-cells are not fully restored in people with HIV (PWH). Moreover, long-lived Th17 cells contribute to HIV persistence under ART. However, the molecular mechanisms underlying these observations remain understudied.
Methods: mRNA-sequencing was performed using Illumina technology on freshly FACS-sorted memory CCR6+CD4+ T-cells from successfully ART-treated (ST), elite controllers (EC), and uninfected donors (HD). Gene expression validation was performed by RT-PCR, flow cytometry, and in vitro functional assays.
Findings: Decreased Th17 cell frequencies in STs and ECs versus HDs coincided with reduced Th17-lineage cytokine production in vitro. Accordingly, the RORγt/RORC2 repressor NR1D1 was upregulated, while the RORγt/RORC2 inducer Semaphorin 4D was decreased in memory CCR6+ T-cells of STs and ECs versus HDs. The presence of HIV-DNA in memory CCR6+ T-cells of ST and EC corresponded with the downregulation of HIV restriction factors (SERINC3, KLF3, and RNF125) and HIV inhibitors (tetraspanins), along with increased expression of the HIV-dependency factor MRE11, indicative of higher susceptibility/permissiveness to HIV-1 infection. Furthermore, markers of DNA damage/modification were elevated in memory CCR6+ T-cells of STs and ECs versus HDs, in line with their increased activation (CD38/HLA-DR), senescence/exhaustion phenotype (CTLA-4/PD-1/CD57) and their decreased expression of proliferation marker Ki-67.
Interpretation: These results reveal new molecular mechanisms of Th17 cell deficit in ST and EC PWH despite a successful control of HIV-1 replication. This knowledge points to potential therapeutic interventions to limit HIV-1 infection and restore frequencies, effector functions, and senescence/exhaustion in Th17 cells.
Funding: This study was funded by the Canadian Institutes of Health Research (CIHR, operating grant MOP 142294, and the Canadian HIV Cure Enterprise [CanCURE 2.0] Team Grant HB2 164064), and in part, by the Réseau SIDA et maladies infectieuses du Fonds de recherche du Québec-Santé (FRQ-S).
Keywords: CCR6; DNA damage; DNA repair; HIV infection; HIV persistence; Th17 cells; Transcriptomics.
Copyright © 2024 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of interests We have no competing interest to declare.
Figures










References
-
- Harrington L.E., Hatton R.D., Mangan P.R., et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6(11):1123–1132. - PubMed
-
- Yero A., Bouassa R.M., Ancuta P., Estaquier J., Jenabian M.A. Immuno-metabolic control of the balance Th17-polarized and regulatory T-cells during HIV infection. Cytokine Growth Factor Rev. 2023;69:1–13. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

