Histone chaperone HIRA, promyelocytic leukemia protein, and p62/SQSTM1 coordinate to regulate inflammation during cell senescence
- PMID: 39178863
- PMCID: PMC11390980
- DOI: 10.1016/j.molcel.2024.08.006
Histone chaperone HIRA, promyelocytic leukemia protein, and p62/SQSTM1 coordinate to regulate inflammation during cell senescence
Abstract
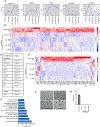
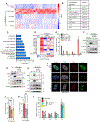
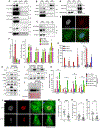
Cellular senescence, a stress-induced stable proliferation arrest associated with an inflammatory senescence-associated secretory phenotype (SASP), is a cause of aging. In senescent cells, cytoplasmic chromatin fragments (CCFs) activate SASP via the anti-viral cGAS/STING pathway. Promyelocytic leukemia (PML) protein organizes PML nuclear bodies (NBs), which are also involved in senescence and anti-viral immunity. The HIRA histone H3.3 chaperone localizes to PML NBs in senescent cells. Here, we show that HIRA and PML are essential for SASP expression, tightly linked to HIRA's localization to PML NBs. Inactivation of HIRA does not directly block expression of nuclear factor κB (NF-κB) target genes. Instead, an H3.3-independent HIRA function activates SASP through a CCF-cGAS-STING-TBK1-NF-κB pathway. HIRA physically interacts with p62/SQSTM1, an autophagy regulator and negative SASP regulator. HIRA and p62 co-localize in PML NBs, linked to their antagonistic regulation of SASP, with PML NBs controlling their spatial configuration. These results outline a role for HIRA and PML in the regulation of SASP.
Keywords: CCF; HIRA; NF-κB pathway; PML; PML NBs; SASP; cGAS-STING signaling; p62/SQSTM; senescence.
Copyright © 2024 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures




Update of
-
Histone chaperone HIRA, Promyelocytic Leukemia (PML) protein and p62/SQSTM1 coordinate to regulate inflammation during cell senescence.bioRxiv [Preprint]. 2024 Jun 25:2023.06.24.546372. doi: 10.1101/2023.06.24.546372. bioRxiv. 2024. Update in: Mol Cell. 2024 Sep 5;84(17):3271-3287.e8. doi: 10.1016/j.molcel.2024.08.006. PMID: 38979156 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

