MEF2B C-terminal mutations enhance transcriptional activity and stability to drive B cell lymphomagenesis
- PMID: 39179580
- PMCID: PMC11343756
- DOI: 10.1038/s41467-024-51644-8
MEF2B C-terminal mutations enhance transcriptional activity and stability to drive B cell lymphomagenesis
Abstract
The myocyte enhancer factor 2B (MEF2B) transcription factor is frequently mutated in germinal center (GC)-derived B-cell lymphomas. Its ammino (N)-terminal mutations drive lymphomagenesis by escaping interaction with transcriptional repressors, while the function of carboxy (C)-terminal mutations remains to be elucidated. Here, we show that MEF2B C-tail is physiologically phosphorylated at specific residues and phosphorylation at serine (S)324 is impaired by lymphoma-associated mutations. Lack of phosphorylation at S324 enhances the interaction of MEF2B with the SWI/SNF chromatin remodeling complex, leading to higher transcriptional activity. In addition, these mutants show an increased protein stability due to impaired interaction with the CUL3/KLHL12 ubiquitin complex. Mice expressing a phosphorylation-deficient lymphoma-associated MEF2B mutant display GC enlargement and develop GC-derived lymphomas, when crossed with Bcl2 transgenic mice. These results unveil converging mechanisms of action for a diverse spectrum of MEF2B mutations, all leading to its dysregulation and GC B-cell lymphomagenesis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


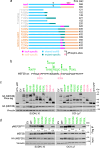

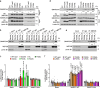




References
-
- Swerdlow SH, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (IARC Publications 2017).
-
- Casulo, C. et al. Early relapse of follicular lymphoma after rituximab plus cyclophosphamide, doxorubicin, vincristine, and prednisone defines patients at high risk for death: an analysis from the National Lymphocare Study. J. Clin. Oncol.33, 2516–2522 (2015). 10.1200/JCO.2014.59.7534 - DOI - PMC - PubMed
-
- Victora, G. D. & Nussenzweig, M. C. Germinal centers. Annu. Rev. Immunol.40, 413–442 (2022). - PubMed
MeSH terms
Substances
Grants and funding
- R35CA-210105/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- P30CA013696/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- S10OD020056/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- R35 CA210105/CA/NCI NIH HHS/United States
- S10 OD020056/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

