The bacterial microbiome and resistome of house dust mites in Irish homes
- PMID: 39179632
- PMCID: PMC11343766
- DOI: 10.1038/s41598-024-70686-y
The bacterial microbiome and resistome of house dust mites in Irish homes
Abstract
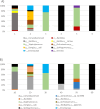
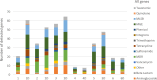
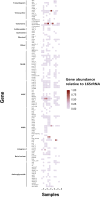
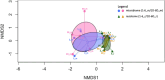
Dust samples were collected from Irish homes. House Dust Mite and storage mites were separated from the dust. The microbiome and resistome of mites and originating dust were assessed using a culture-independent approach. The bacterial microbiome of mites and dust were predominantly populated by Staphylococci. There was a highly significant (P = 0.005; Spearman's rank test) correlation between the bacterial microbiome of mites and the dust. One-hundred and eighteen antimicrobial resistance genes (ARGs) were associated with mites and 176 with dust. Both contained ARGs encoding resistance for multi drug resistances, macrolide-lincosamide-streptogramin B, mobile genetic elements, Beta-lactam, Tetracycline and Aminoglycosides. By contrast, 15 ARGs were found for a laboratory-grown strain of Dermatophagoides pteronyssinus. A significant difference (P = 0.03; t test) was found in means between the resistome of mites and the household dust from which they emanated. No significant correlations (P = 0.23 and P = 0.22; Mantel test) were observed between the microbiome and resistome of mite and dust samples. There was not a significant difference (P = 0.54; t-test) between the means of ARGs for homes with and without a history of antibiotic use.
Keywords: 16 S rRNA microbiome; Antibiotic resistant genes; Dust mites; Dust samples; Resistome; qPCR SmartChip.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Brasche, S. & Bischof, W. Daily time spent indoors in German homes—Baseline data for the assessment of indoor exposure of German occupants. Int. J. Hyg. Environ. Health208, 247–253 (2005). - PubMed
-
- Morawska, L. et al. Airborne particles in indoor environment of homes, schools, offices and aged care facilities: The main routes of exposure. Environ. Int.108, 75–83. 10.1016/j.envint.2017.07.025 (2017). - PubMed
-
- Viegas, C. et al. Settled dust assessment in clinical environment: Useful for the evaluation of a wider bioburden spectrum. Taylor Fr.31, 160–178 (2021). - PubMed
-
- Melymuk, L., Demirtepe, H. & Jílková, S. R. Indoor dust and associated chemical exposures. Curr. Opin. Environ. Sci. Health15, 1–6 (2020).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

