Improved pokeweed genome assembly and early gene expression changes in response to jasmonic acid
- PMID: 39179987
- PMCID: PMC11344361
- DOI: 10.1186/s12870-024-05446-1
Improved pokeweed genome assembly and early gene expression changes in response to jasmonic acid
Abstract
Background: Jasmonic acid (JA) is a phytohormone involved in regulating responses to biotic and abiotic stress. Although the JA pathway is well characterized in model plants such as Arabidopsis thaliana, less is known about many non-model plants. Phytolacca americana (pokeweed) is native to eastern North Americana and is resilient to environmental stress. The goal of this study was to produce a publicly available pokeweed genome assembly and annotations and use this resource to determine how early response to JA changes gene expression, with particular focus on genes involved in defense.
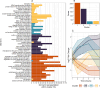
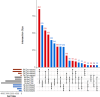
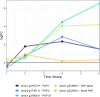
Results: We assembled the pokeweed genome de novo from approximately 30 Gb of PacBio Hifi long reads and achieved an NG50 of ~ 13.2 Mb and a minimum 93.9% complete BUSCO score for gene annotations. With this reference, we investigated the early changes in pokeweed gene expression following JA treatment. Approximately 5,100 genes were differentially expressed during the 0-6 h time course with almost equal number of genes with increased and decreased transcript levels. Cluster and gene ontology analyses indicated the downregulation of genes associated with photosynthesis and upregulation of genes involved in hormone signaling and defense. We identified orthologues of key transcription factors and constructed the first JA gene response network integrated with our transcriptomic data from orthologues of Arabidopsis genes. We discovered that pokeweed did not use leaf senescence as a means of reallocating resources during stress; rather, most secondary metabolite synthesis genes were constitutively expressed, suggesting that pokeweed directs its resources for survival over the long term. In addition, pokeweed synthesizes several RNA N-glycosylases hypothesized to function in defense, each with unique expression profiles in response to JA.
Conclusions: Our investigation of the early response of pokeweed to JA illustrates patterns of gene expression involved in defence and stress tolerance. Pokeweed provides insight into the defense mechanisms of plants beyond those observed in research models and crops, and further study may yield novel approaches to improving the resilience of plants to environmental changes. Our assembled pokeweed genome is the first within the taxonomic family Phytolaccaceae to be publicly available for continued research.
Keywords: Phytolacca americana; Gene regulatory network; Genome assembly; Jasmonic acid; Pokeweed antiviral protein; RNA-seq; Stress response.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

