Expression levels of core spliceosomal proteins modulate the MBNL-mediated spliceopathy in DM1
- PMID: 39180495
- PMCID: PMC11540926
- DOI: 10.1093/hmg/ddae125
Expression levels of core spliceosomal proteins modulate the MBNL-mediated spliceopathy in DM1
Abstract
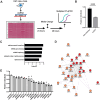
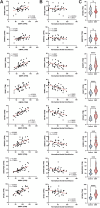
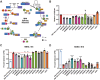
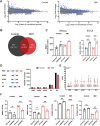
Myotonic dystrophy type 1 (DM1) is a heterogeneous multisystemic disease caused by a CTG repeat expansion in DMPK. Transcription of the expanded allele produces toxic CUG repeat RNA that sequesters the MBNL family of alternative splicing (AS) regulators into ribonuclear foci, leading to pathogenic mis-splicing. To identify genetic modifiers of toxic CUG RNA levels and the spliceopathy, we performed a genome-scale siRNA screen using an established HeLa DM1 repeat-selective screening platform. We unexpectedly identified core spliceosomal proteins as a new class of modifiers that rescue the spliceopathy in DM1. Modest knockdown of one of our top hits, SNRPD2, in DM1 fibroblasts and myoblasts, significantly reduces DMPK expression and partially rescues MBNL-regulated AS dysfunction. While the focus on the DM1 spliceopathy has centered around the MBNL proteins, our work reveals an unappreciated role for MBNL:spliceosomal protein stoichiometry in modulating the spliceopathy, revealing new biological and therapeutic avenues for DM1.
Keywords: alternative splicing; genetic modifiers; genome screen; myotonic dystrophy; repeat expansion diseases.
© The Author(s) 2024. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures


References
-
- Brook JD, McCurrach ME, Harley HG. et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell 1992;68:799–808. - PubMed
-
- Harley HG, Brook JD, Rundle SA. et al. Expansion of an unstable DNA region and phenotypic variation in myotonic dystrophy. Nature 1992;355:545–546. - PubMed
-
- Mahadevan M, Tsilfidis C, Sabourin L. et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science 1992;255:1253–1255. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

