Salmonella re-engineers the intestinal environment to break colonization resistance in the presence of a compositionally intact microbiota
- PMID: 39181125
- PMCID: PMC11466686
- DOI: 10.1016/j.chom.2024.07.025
Salmonella re-engineers the intestinal environment to break colonization resistance in the presence of a compositionally intact microbiota
Abstract
The gut microbiota prevents harmful microbes from entering the body, a function known as colonization resistance. The enteric pathogen Salmonella enterica serovar (S.) Typhimurium uses its virulence factors to break colonization resistance through unknown mechanisms. Using metabolite profiling and genetic analysis, we show that the initial rise in luminal pathogen abundance was powered by a combination of aerobic respiration and mixed acid fermentation of simple sugars, such as glucose, which resulted in their depletion from the metabolome. The initial rise in the abundance of the pathogen in the feces coincided with a reduction in the cecal concentrations of acetate and butyrate and an increase in epithelial oxygenation. Notably, these changes in the host environment preceded changes in the microbiota composition. We conclude that changes in the host environment can weaken colonization resistance even in the absence of overt compositional changes in the gut microbiota.
Keywords: Salmonella; colonization resistance; microbiota; mixed acid fermentation; short-chain fatty acids.
Copyright © 2024 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



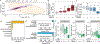
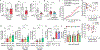

References
MeSH terms
Substances
Grants and funding
- R56 AI112949/AI/NIAID NIH HHS/United States
- T32 AI060555/AI/NIAID NIH HHS/United States
- R01 AI044170/AI/NIAID NIH HHS/United States
- R21 AI146432/AI/NIAID NIH HHS/United States
- R01 AI096528/AI/NIAID NIH HHS/United States
- R37 AI044170/AI/NIAID NIH HHS/United States
- R21 AI153069/AI/NIAID NIH HHS/United States
- R01 AI112445/AI/NIAID NIH HHS/United States
- S10 OD010786/OD/NIH HHS/United States
- KL2 TR001859/TR/NCATS NIH HHS/United States
- UL1 TR001860/TR/NCATS NIH HHS/United States
- R01 AI112949/AI/NIAID NIH HHS/United States
- R01 DK138912/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

