Single Acetylation-mimetic Mutation in TDP-43 Nuclear Localization Signal Disrupts Importin α1/β Signaling
- PMID: 39181183
- PMCID: PMC11443512
- DOI: 10.1016/j.jmb.2024.168751
Single Acetylation-mimetic Mutation in TDP-43 Nuclear Localization Signal Disrupts Importin α1/β Signaling
Abstract
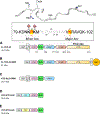
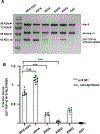
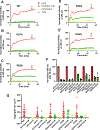
Cytoplasmic aggregation of the TAR-DNA binding protein of 43 kDa (TDP-43) is the hallmark of sporadic amyotrophic lateral sclerosis (ALS). Most ALS patients with TDP-43 aggregates in neurons and glia do not have mutations in the TDP-43 gene but contain aberrantly post-translationally modified TDP-43. Here, we found that a single acetylation-mimetic mutation (K82Q) near the TDP-43 minor Nuclear Localization Signal (NLS) box, which mimics a post-translational modification identified in an ALS patient, can lead to TDP-43 mislocalization to the cytoplasm and irreversible aggregation. We demonstrate that the acetylation mimetic disrupts binding to importins, halting nuclear import and preventing importin α1/β anti-aggregation activity. We propose that perturbations near the NLS are an additional mechanism by which a cellular insult other than a genetically inherited mutation leads to TDP-43 aggregation and loss of function. Our findings are relevant to deciphering the molecular etiology of sporadic ALS.
Keywords: ALS; TDP-43; importin α1; importin β; nuclear localization signal.
Copyright © 2024 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- Andersen PM, Al-Chalabi A. Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7:603–15. - PubMed
-
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

