METI: deep profiling of tumor ecosystems by integrating cell morphology and spatial transcriptomics
- PMID: 39181865
- PMCID: PMC11344794
- DOI: 10.1038/s41467-024-51708-9
METI: deep profiling of tumor ecosystems by integrating cell morphology and spatial transcriptomics
Abstract
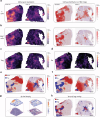
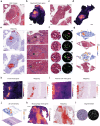
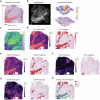
Recent advances in spatial transcriptomics (ST) techniques provide valuable insights into cellular interactions within the tumor microenvironment (TME). However, most analytical tools lack consideration of histological features and rely on matched single-cell RNA sequencing data, limiting their effectiveness in TME studies. To address this, we introduce the Morphology-Enhanced Spatial Transcriptome Analysis Integrator (METI), an end-to-end framework that maps cancer cells and TME components, stratifies cell types and states, and analyzes cell co-localization. By integrating spatial transcriptomics, cell morphology, and curated gene signatures, METI enhances our understanding of the molecular landscape and cellular interactions within the tissue. We evaluate the performance of METI on ST data generated from various tumor tissues, including gastric, lung, and bladder cancers, as well as premalignant tissues. We also conduct a quantitative comparison of METI with existing clustering and cell deconvolution tools, demonstrating METI's robust and consistent performance.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

