The Chlamydia pneumoniae effector SemD exploits its host's endocytic machinery by structural and functional mimicry
- PMID: 39181890
- PMCID: PMC11344771
- DOI: 10.1038/s41467-024-51681-3
The Chlamydia pneumoniae effector SemD exploits its host's endocytic machinery by structural and functional mimicry
Abstract
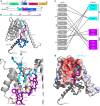
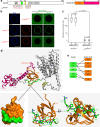
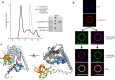
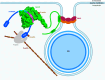
To enter epithelial cells, the obligate intracellular pathogen Chlamydia pneumoniae secretes early effector proteins, which bind to and modulate the host-cell's plasma membrane and recruit several pivotal endocytic host proteins. Here, we present the high-resolution structure of an entry-related chlamydial effector protein, SemD. Co-crystallisation of SemD with its host binding partners demonstrates that SemD co-opts the Cdc42 binding site to activate the actin cytoskeleton regulator N-WASP, making active, GTP-bound Cdc42 superfluous. While SemD binds N-WASP much more strongly than Cdc42 does, it does not bind the Cdc42 effector protein FMNL2, indicating effector protein specificity. Furthermore, by identifying flexible and structured domains, we show that SemD can simultaneously interact with the membrane, the endocytic protein SNX9, and N-WASP. Here, we show at the structural level how a single effector protein can hijack central components of the host's endocytic system for efficient internalization.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

