Environmental community transcriptomics: strategies and struggles
- PMID: 39183066
- PMCID: PMC11735753
- DOI: 10.1093/bfgp/elae033
Environmental community transcriptomics: strategies and struggles
Abstract
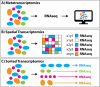
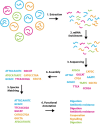
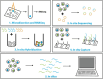
Transcriptomics is the study of RNA transcripts, the portion of the genome that is transcribed, in a specific cell, tissue, or organism. Transcriptomics provides insight into gene expression patterns, regulation, and the underlying mechanisms of cellular processes. Community transcriptomics takes this a step further by studying the RNA transcripts from environmental assemblies of organisms, with the intention of better understanding the interactions between members of the community. Community transcriptomics requires successful extraction of RNA from a diverse set of organisms and subsequent analysis via mapping those reads to a reference genome or de novo assembly of the reads. Both, extraction protocols and the analysis steps can pose hurdles for community transcriptomics. This review covers advances in transcriptomic techniques and assesses the viability of applying them to community transcriptomics.
Keywords: label-free cell sorting; metatranscriptomics; single cell transcriptomics; spatial transcriptomics; transcriptomics.
Published by Oxford University Press 2024.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

