Multi-omics integration reveals the oncogenic role of eccDNAs in diffuse large B-cell lymphoma through STING signalling
- PMID: 39183480
- PMCID: PMC11345442
- DOI: 10.1002/ctm2.1815
Multi-omics integration reveals the oncogenic role of eccDNAs in diffuse large B-cell lymphoma through STING signalling
Erratum in
-
Erratum for the Clinical and Translational Medicine "Multi-omics integration reveals the oncogenic role of eccDNAs in diffuse large B-cell lymphoma through STING signalling" by Zijuan Wu et al.Clin Transl Med. 2025 Apr;15(4):e70301. doi: 10.1002/ctm2.70301. Clin Transl Med. 2025. PMID: 40170263 Free PMC article. No abstract available.
-
Erratum for the Clinical and Translational Medicine "Multi-omics integration reveals the oncogenic role of eccDNAs in diffuse large B-cell lymphoma through STING signalling" by Zijuan Wu et al.Clin Transl Med. 2025 Jul;15(7):e70312. doi: 10.1002/ctm2.70312. Clin Transl Med. 2025. PMID: 40708210 Free PMC article. No abstract available.
Abstract
Background: Extrachromosomal circular DNAs (eccDNAs), a type of double-stranded DNAs (dsDNAs) that facilitate the activation of the DNA sensing machinery, have been implicated in the progression and prognosis of various diseases. While the roles of eccDNAs remain contentious, their significance in diffuse large B-cell lymphoma (DLBCL) has not been reported.
Methods: Circular DNA sequencing (circle-seq) was used to demonstrate the expression profile of eccDNAs in DLBCL, and atomic force microscopy to validate the presence of eccDNAs. CCK-8 and scRNA-seq techniques were employed to uncover the activation of eccDNA in the STING pathway, leading to enhanced cell proliferation. Chemotherapeutic drugs were used to test the hypothesis that DNA damage induces the production of eccDNA, thereby activating the STING pathway independent of cGAS. GEO databases were used for verification of the prognosis of the eccDNA-related genes, and animal models were used to investigate the synergistic effects of DNA damage therapy in combination with STING inhibitors on anti-tumour responses.
Results: EccDNAs were widely expressed in DLBCL and associated with the prognosis of patients. Elevated abundance of eccDNAs promoted the progression of DLBCL. Chemotherapeutic drugs-induced DNA damage triggered the generation of eccDNAs, resulting in the activation of the STING signalling in a cGAS-independent manner. Moreover, inhibition of STING exerted a synergistic anti-tumour effect with cisplatin.
Conclusions: EccDNAs induced by DNA damage exert an oncogenic role in DLBCL via activating the STING signalling independently of cGAS. This finding offers a rational therapeutic strategy combining chemotherapy with targeting STING.
Highlights: EccDNAs induced by DNA damage exert an oncogenic role in DLBCL via activating the STING signalling independently of cGAS. The combined treatment of chemotherapeutic drugs with STING inhibitor significantly delayed the tumor progression, providing new insights into the therapeutic strategy for patients with DLBCL, particularly the relapsed and/or refractory (R/R) ones.
Keywords: DNA damage; Diffuse large B‐cell lymphoma; EccDNAs; STING signalling.
© 2024 The Author(s). Clinical and Translational Medicine published by John Wiley & Sons Australia, Ltd on behalf of Shanghai Institute of Clinical Bioinformatics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



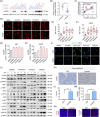
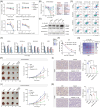
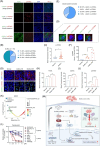
References
MeSH terms
Substances
Grants and funding
- 82200213/National Natural Science Foundation of China
- 82170185/National Natural Science Foundation of China
- 82370193/National Natural Science Foundation of China
- 81570184/National Natural Science Foundation of China
- BK20211376/Natural Science Foundation of Jiangsu Province
- BE2023775/Jiangsu Province Science and Technology plan
- KYCX24_2038/Postgraduate Research & Practice Innovation Program of Jiangsu Province
- CZ0121002010037/Jiangsu Province Hospital High-level Talent Cultivation Program (Phase I)
- ZDXK202209/Jiangsu Province Capability Improvement Project through Science, Technology and Education
LinkOut - more resources
Full Text Sources
Research Materials
