Disruption of the Uty epigenetic regulator locus in hematopoietic cells phenocopies the profibrotic attributes of Y chromosome loss in heart failure
- PMID: 39183958
- PMCID: PMC11343478
- DOI: 10.1038/s44161-024-00441-z
Disruption of the Uty epigenetic regulator locus in hematopoietic cells phenocopies the profibrotic attributes of Y chromosome loss in heart failure
Abstract
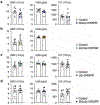
Heart failure affects millions of people worldwide, with men exhibiting a higher incidence than women. Our previous work has shown that mosaic loss of the Y chromosome (LOY) in leukocytes is causally associated with an increased risk for heart failure. Here, we show that LOY macrophages from the failing hearts of humans with dilated cardiomyopathy exhibit widespread changes in gene expression that correlate with cardiac fibroblast activation. Moreover, we identify the ubiquitously transcribed t et ratricopeptide Y-linked (Uty) gene in leukocytes as a causal locus for an accelerated progression of heart failure in male mice with LOY. We demonstrate that Uty disruption leads to epigenetic alterations in both monocytes and macrophages, increasing the propensity of differentiation into profibrotic macrophages. Treatment with a transforming growth factor-β-neutralizing antibody prevented the cardiac pathology associated with Uty deficiency in leukocytes. These findings shed light on the mechanisms that contribute to the higher incidence of heart failure in men.
Conflict of interest statement
Competing interests The authors declare no competing interests.
Figures

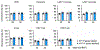










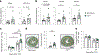

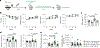
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
