Identification of FDA-approved drugs that induce heart regeneration in mammals
- PMID: 39183959
- PMCID: PMC11343477
- DOI: 10.1038/s44161-024-00450-y
Identification of FDA-approved drugs that induce heart regeneration in mammals
Abstract
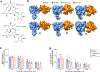
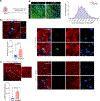
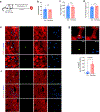
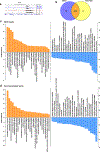
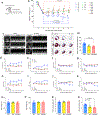
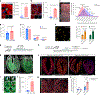
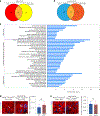
Targeting Meis1 and Hoxb13 transcriptional activity could be a viable therapeutic strategy for heart regeneration. In this study, we performd an in silico screening to identify FDA-approved drugs that can inhibit Meis1 and Hoxb13 transcriptional activity based on the resolved crystal structure of Meis1 and Hoxb13 bound to DNA. Paromomycin (Paro) and neomycin (Neo) induced proliferation of neonatal rat ventricular myocytes in vitro and displayed dose-dependent inhibition of Meis1 and Hoxb13 transcriptional activity by luciferase assay and disruption of DNA binding by electromobility shift assay. X-ray crystal structure revealed that both Paro and Neo bind to Meis1 near the Hoxb13-interacting domain. Administration of Paro-Neo combination in adult mice and in pigs after cardiac ischemia/reperfusion injury induced cardiomyocyte proliferation, improved left ventricular systolic function and decreased scar formation. Collectively, we identified FDA-approved drugs with therapeutic potential for induction of heart regeneration in mammals.
Conflict of interest statement
Competing interests M.G. is founder, consultant, member of the board and equity holder in Forcefield Therapeutics, Heqet Therapeutics and Purespring Therapeutics. All other authors declare no competing interests.
Figures










Comment in
-
Toward drug-induced heart regeneration.Nat Cardiovasc Res. 2024 Mar;3(3):260-261. doi: 10.1038/s44161-024-00446-8. Nat Cardiovasc Res. 2024. PMID: 39196122 No abstract available.
References
-
- Poss KD Getting to the heart of regeneration in zebrafish. Semin. Cell Dev. Biol. 18, 36–45 (2007). - PubMed
-
- Walsh S, Ponten A, Fleischmann BK & Jovinge S Cardiomyocyte cell cycle control and growth estimation in vivo—an analysis based on cardiomyocyte nuclei. Cardiovasc. Res. 86, 365–373 (2010). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
