Purinergic exposure induces epigenomic and transcriptomic-mediated preconditioning resembling epilepsy-associated microglial states
- PMID: 39184445
- PMCID: PMC11342283
- DOI: 10.1016/j.isci.2024.110546
Purinergic exposure induces epigenomic and transcriptomic-mediated preconditioning resembling epilepsy-associated microglial states
Abstract
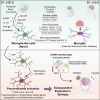
Microglia play a crucial role in a range of neuropathologies through exacerbated activation. Microglial inflammatory responses can be influenced by prior exposures to noxious stimuli, like increased levels of extracellular adenosine and ATP. These are characteristic of brain insults like epileptic seizures and could potentially shape subsequent responses through epigenetic regulation. We investigated DNA methylation and expression changes in human microglia-like cells differentiated from monocytes following ATP-mediated preconditioning. We demonstrate that microglia-like cells display homeostatic microglial features, shown by surface markers, transcriptome, and DNA methylome. After exposure to ATP, TLR-mediated activation leads to an exacerbated pro-inflammatory response. These changes are accompanied by methylation and transcriptional reprogramming associated with enhanced immune-related functions. The reprogramming associated with ATP-mediated preconditioning leads to profiles found in microglial subsets linked to epilepsy. Purine-driven microglia immune preconditioning drives epigenetic and transcriptional changes that could contribute to altered functions of microglia during seizure development and progression.
Keywords: Epigenetics; Epilepsy; Microglia; Purines; Transcriptomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Zhan Y., Paolicelli R.C., Sforazzini F., Weinhard L., Bolasco G., Pagani F., Vyssotski A.L., Bifone A., Gozzi A., Ragozzino D., Gross C.T. Deficient neuron-microglia signaling results in impaired functional brain connectivity and social behavior. Nat. Neurosci. 2014;17:400–406. doi: 10.1038/nn.3641. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

