Complete mitochondrial genome assembly of Zizania latifolia and comparative genome analysis
- PMID: 39184575
- PMCID: PMC11341417
- DOI: 10.3389/fpls.2024.1381089
Complete mitochondrial genome assembly of Zizania latifolia and comparative genome analysis
Abstract
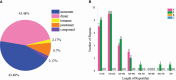
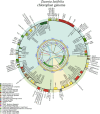
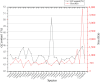
Zizania latifolia (Griseb.) Turcz. ex Stapf has been cultivated as a popular aquatic vegetable in China due to its important nutritional, medicinal, ecological, and economic values. The complete mitochondrial genome (mitogenome) of Z. latifolia has not been previously studied and reported, which has hindered its molecular systematics and understanding of evolutionary processes. Here, we assembled the complete mitogenome of Z. latifolia and performed a comprehensive analysis including genome organization, repetitive sequences, RNA editing event, intercellular gene transfer, phylogenetic analysis, and comparative mitogenome analysis. The mitogenome of Z. latifolia was estimated to have a circular molecule of 392,219 bp and 58 genes consisting of three rRNA genes, 20 tRNA genes, and 35 protein-coding genes (PCGs). There were 46 and 20 simple sequence repeats (SSRs) with different motifs identified from the mitogenome and chloroplast genome of Z. latifolia, respectively. Furthermore, 49 homologous fragments were observed to transfer from the chloroplast genome to the mitogenome of Z. latifolia, accounting for 47,500 bp, presenting 12.1% of the whole mitogenome. In addition, there were 11 gene-containing homologous regions between the mitogenome and chloroplast genome of Z. latifolia. Also, approximately 85% of fragments from the mitogenome were duplicated in the Z. latifolia nuclear genome. Selection pressure analysis revealed that most of the mitochondrial genes were highly conserved except for ccmFc, ccmFn, matR, rps1, and rps3. A total of 93 RNA editing sites were found in the PCGs of the mitogenome. Z. latifolia and Oryza minuta are the most closely related, as shown by collinear analysis and the phylogenetic analysis. We found that repeat sequences and foreign sequences in the mitogenomes of Oryzoideae plants were associated with genome rearrangements. In general, the availability of the Z. latifolia mitogenome will contribute valuable information to our understanding of the molecular and genomic aspects of Zizania.
Keywords: Oryzoideae; RNA editing; Zizania latifolia; mitochondrial genome; phylogenetic; repeat sequence; synteny.
Copyright © 2024 Luo, Gu, Gao, Li, Zhang and Zhu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Beser N., Mutafcilar Z. C., Hasancebi S. (2021). Diversity analysis of the rice cultivars (Oryza sativa L.) by utilizing SSRs rice diversity by SSRs. J. Food Process. AND PRESERVATION 45 (2). doi: 10.1111/jfpp.15232 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous

