This is a preprint.
Identification of biomarkers for COVID-19 associated secondary hemophagocytic lymphohistiocytosis
- PMID: 39185173
- PMCID: PMC11343163
- DOI: 10.1101/2024.08.13.607855
Identification of biomarkers for COVID-19 associated secondary hemophagocytic lymphohistiocytosis
Update in
-
Proteomic Analyses in COVID-19-Associated Secondary Hemophagocytic Lymphohistiocytosis.Crit Care Explor. 2025 Jan 31;7(2):e1203. doi: 10.1097/CCE.0000000000001203. eCollection 2025 Feb 1. Crit Care Explor. 2025. PMID: 39888602 Free PMC article.
Abstract
Objectives: We aimed to define and validate novel biomarkers that could identify individuals with COVID-19 associated secondary hemophagocytic lymphohistiocytosis (sHLH) and to test whether fatalities due to COVID-19 in the presence of sHLH were associated with specific defects in the immune system.
Design: In two cohorts of adult patients presenting with COVID-19 in 2020 and 2021, clinical lab values and serum proteomics were assessed. Subjects identified as having sHLH were compared to those with COVID-19 without sHLH. Eight deceased patients defined as COVID-sHLH underwent genomic sequencing in order to identify variants in immune-related genes.
Setting: Two tertiary care hospitals in Seattle, Washington (Virginia Mason Medical Center and Harborview Medical Center).
Patients: 186 patients with COVID-19.
Interventions: None.
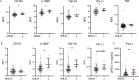
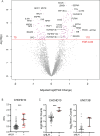
Measurements and main results: Nine percent of enrolled COVID-19 subjects met our defined criteria for sHLH. Using broad serum proteomic approaches (O-link and SomaScan), we identified three biomarkers for COVID-19 associated sHLH (soluble PD-L1, TNF-R1, and IL-18BP), supporting a role for proteins previously associated with other forms of sHLH (IL-18BP and sTNF-R1). We also identified novel biomarkers and pathways of COVID-sHLH, including sPD-L1 and the syntaxin pathway. We detected variants in several genes involved in immune responses in individuals with COVID-sHLH, including in DOCK8 and in TMPRSS15, suggesting that genetic alterations in immune-related genes may contribute to hyperinflammation and fatal outcomes in COVID-19.
Conclusions: Biomarkers of COVID-19 associated sHLH, such as soluble PD-L1, and pathways, such as the syntaxin pathway, and variants in immune genes in these individuals, suggest critical roles for the immune response in driving sHLH in the context of COVID-19.
Figures
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
