This is a preprint.
Experimental and Computational Methods for Allelic Imbalance Analysis from Single-Nucleus RNA-seq Data
- PMID: 39185246
- PMCID: PMC11343128
- DOI: 10.1101/2024.08.13.607784
Experimental and Computational Methods for Allelic Imbalance Analysis from Single-Nucleus RNA-seq Data
Abstract
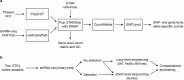
Single-cell RNA-seq (scRNA-seq) is emerging as a powerful tool for understanding gene function across diverse cells. Recently, this has included the use of allele-specific expression (ASE) analysis to better understand how variation in the human genome affects RNA expression at the single-cell level. We reasoned that because intronic reads are more prevalent in single-nucleus RNA-Seq (snRNA-Seq), and introns are under lower purifying selection and thus enriched for genetic variants, that snRNA-seq should facilitate single-cell analysis of ASE. Here we demonstrate how experimental and computational choices can improve the results of allelic imbalance analysis. We explore how experimental choices, such as RNA source, read length, sequencing depth, genotyping, etc., impact the power of ASE-based methods. We developed a new suite of computational tools to process and analyze scRNA-seq and snRNA-seq for ASE. As hypothesized, we extracted more ASE information from reads in intronic regions than those in exonic regions and show how read length can be set to increase power. Additionally, hybrid selection improved our power to detect allelic imbalance in genes of interest. We also explored methods to recover allele-specific isoform expression levels from both long- and short-read snRNA-seq. To further investigate ASE in the context of human disease, we applied our methods to a Parkinson's disease cohort of 94 individuals and show that ASE analysis had more power than eQTL analysis to identify significant SNP/gene pairs in our direct comparison of the two methods. Overall, we provide an end-to-end experimental and computational approach for future studies.
Keywords: Parkinson’s disease; RNA-seq; Single-cell; allele-specific expression; variant to function.
Conflict of interest statement
Declarations Competing Interests A.M.A., K.G., and J.T.S. are inventors on a licensed, pending international patent application, having serial number PCT/US2021/037226, filed by Broad Institute of MIT and Havard, Massachusetts General Hospital and Massachusetts Institute of Technology, directed to certain subject matter related to the MAS-seq/Kinnex method described in this manuscript. From October 19, 2020, O.R.R. is an employee of Genentech and has equity in Roche. O.R.R. is a co-inventor on patent applications filed by the Broad Institute for inventions related to single-cell genomics. She has given numerous lectures on the subject of single-cell genomics to a wide variety of audiences and, in some cases, has received remuneration to cover time and costs. A.R. is a cofounder of and equity holder in Celsius Therapeutics, an equity holder in Immunitas and was a Scientific Advisory Board member of Thermo Fisher Scientific, Syros Pharmaceuticals, Neogene Therapeutics and Asimov until 31 July 2020. A.R. has been an employee of Genentech (member of the Roche Group) since August 2020 and has equity in Roche. The other authors declare that they have no competing interests.
Figures




References
-
- Oelen R, de Vries DH, Brugge H, Gordon MG, Vochteloo M, single-cell e Qc, Consortium B, Ye CJ, Westra HJ, Franke L, van der Wijst MGP: Single-cell RNA-sequencing of peripheral blood mononuclear cells reveals widespread, context-specific gene expression regulation upon pathogenic exposure. Nat Commun 2022, 13:3267. - PMC - PubMed
-
- Yazar S, Alquicira-Hernandez J, Wing K, Senabouth A, Gordon MG, Andersen S, Lu Q, Rowson A, Taylor TRP, Clarke L, et al. : Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 2022, 376:eabf3041. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
