Modeling APOE ε4 familial Alzheimer's disease in directly converted 3D brain organoids
- PMID: 39185458
- PMCID: PMC11341472
- DOI: 10.3389/fnagi.2024.1435445
Modeling APOE ε4 familial Alzheimer's disease in directly converted 3D brain organoids
Abstract
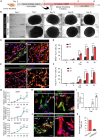
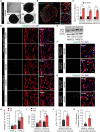
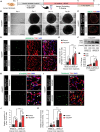
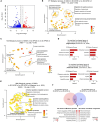
Brain organoids have become a valuable tool for studying human brain development, disease modeling, and drug testing. However, generating brain organoids with mature neurons is time-intensive and often incomplete, limiting their utility in studying age-related neurodegenerative diseases such as Alzheimer's disease (AD). Here, we report the generation of 3D brain organoids from human fibroblasts through direct reprogramming, with simplicity, efficiency, and reduced variability. We also demonstrate that induced brain organoids from APOE ε4 AD patient fibroblasts capture some disease-specific features and pathologies associated with APOE ε4 AD. Moreover, APOE ε4-induced brain organoids with mutant APP overexpression faithfully recapitulate the acceleration of AD-related pathologies, providing a more physiologically relevant and patient-specific model of familial AD. Importantly, transcriptome analysis reveals that gene sets specific to APOE ε4 patient-induced brain organoids are highly similar to those of APOE ε4 post-mortem AD brains. Overall, induced brain organoids from direct reprogramming offer a promising approach for more efficient and controlled studies of neurodegenerative disease modeling.
Keywords: 3D modeling; Alzheimer’s disease; amyloid-beta; apolipoprotein E; direct conversion.
Copyright © 2024 Kim, Kim, Cho, An, Kang, Kim and Kim.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
LinkOut - more resources
Full Text Sources
Miscellaneous

