Prevotella are major contributors of sialidases in the human vaginal microbiome
- PMID: 39186657
- PMCID: PMC11388281
- DOI: 10.1073/pnas.2400341121
Prevotella are major contributors of sialidases in the human vaginal microbiome
Abstract
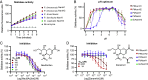
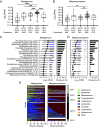
Elevated bacterial sialidase activity in the female genital tract is strongly associated with poor health outcomes including preterm birth and bacterial vaginosis (BV). These negative effects may arise from sialidase-mediated degradation of the protective mucus layer in the cervicovaginal environment. Prior biochemical studies of vaginal bacterial sialidases have focused solely on the BV-associated organism Gardnerella vaginalis. Despite their implications for sexual and reproductive health, sialidases from other vaginal bacteria have not been characterized. Here, we show that vaginal Prevotella species produce sialidases that possess variable activity toward mucin substrates. The sequences of sialidase genes and their presence are largely conserved across clades of Prevotella from different geographies, hinting at their importance globally. Finally, we find that Prevotella sialidase genes and transcripts, including those encoding mucin-degrading sialidases from Prevotella timonensis, are highly prevalent and abundant in human vaginal genomes and transcriptomes. Together, our results identify Prevotella as a critical source of sialidases in the vaginal microbiome, improving our understanding of this detrimental bacterial activity.
Keywords: Prevotella; bacterial vaginosis; mucin; sialidase; vaginal microbiome.
Conflict of interest statement
Competing interests statement:J.R. is a cofounder of LUCA Biologics, a biotechnology company focusing on translating microbiome research into live biotherapeutic drugs for women’s health. D.S.K. serves as equity holder of Day Zero Diagnostics.
Figures



References
-
- Cauci S., McGregor J., Thorsen P., Grove J., Guaschino S., Combination of vaginal pH with vaginal sialidase and prolidase activities for prediction of low birth weight and preterm birth. Am. J. Obstet. Gynecol. 192, 489–496 (2005). - PubMed
-
- Cauci S., Culhane J. F., High sialidase levels increase preterm birth risk among women who are bacterial vaginosis-positive in early gestation. Am. J. Obstet. Gynecol. 204, e1–142.e9 (2011). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

