Fatty acid metabolism constrains Th9 cell differentiation and antitumor immunity via the modulation of retinoic acid receptor signaling
- PMID: 39187636
- PMCID: PMC11528006
- DOI: 10.1038/s41423-024-01209-y
Fatty acid metabolism constrains Th9 cell differentiation and antitumor immunity via the modulation of retinoic acid receptor signaling
Erratum in
-
Author Correction: Fatty acid metabolism constrains Th9 cell differentiation and antitumor immunity via the modulation of retinoic acid receptor signaling.Cell Mol Immunol. 2024 Nov;21(11):1350. doi: 10.1038/s41423-024-01223-0. Cell Mol Immunol. 2024. PMID: 39379605 Free PMC article. No abstract available.
Abstract
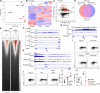
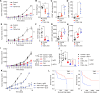
T helper 9 (Th9) cells are interleukin 9 (IL-9)-producing cells that have diverse functions ranging from antitumor immune responses to allergic inflammation. Th9 cells differentiate from naïve CD4+ T cells in the presence of IL-4 and transforming growth factor-beta (TGF-β); however, our understanding of the molecular basis of their differentiation remains incomplete. Previously, we reported that the differentiation of another subset of TGF-β-driven T helper cells, Th17 cells, is highly dependent on de novo lipid biosynthesis. On the basis of these findings, we hypothesized that lipid metabolism may also be important for Th9 cell differentiation. We therefore investigated the differentiation and function of mouse and human Th9 cells in vitro under conditions of pharmacologically or genetically induced deficiency of the intracellular fatty acid content and in vivo in mice genetically deficient in acetyl-CoA carboxylase 1 (ACC1), an important enzyme for fatty acid biosynthesis. Both the inhibition of de novo fatty acid biosynthesis and the deprivation of environmental lipids augmented differentiation and IL-9 production in mouse and human Th9 cells. Mechanistic studies revealed that the increase in Th9 cell differentiation was mediated by the retinoic acid receptor and the TGF-β-SMAD signaling pathways. Upon adoptive transfer, ACC1-inhibited Th9 cells suppressed tumor growth in murine models of melanoma and adenocarcinoma. Together, our findings highlight a novel role of fatty acid metabolism in controlling the differentiation and in vivo functions of Th9 cells.
Keywords: RARα; Th9 cell; anti-tumor effect; fatty acid; immunometabolism; omics analysis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Angkasekwinai P, Dong C. IL-9-producing T cells: potential players in allergy and cancer. Nat Rev Immunol. 2021;21:37–48. 10.1038/s41577-020-0396-0. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

