Robust genome and cell engineering via in vitro and in situ circularized RNAs
- PMID: 39187662
- PMCID: PMC12186994
- DOI: 10.1038/s41551-024-01245-z
Robust genome and cell engineering via in vitro and in situ circularized RNAs
Abstract
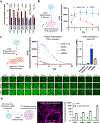
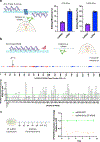
Circularization can improve RNA persistence, yet simple and scalable approaches to achieve this are lacking. Here we report two methods that facilitate the pursuit of circular RNAs (cRNAs): cRNAs developed via in vitro circularization using group II introns, and cRNAs developed via in-cell circularization by the ubiquitously expressed RtcB protein. We also report simple purification protocols that enable high cRNA yields (40-75%) while maintaining low immune responses. These methods and protocols facilitate a broad range of applications in stem cell engineering as well as robust genome and epigenome targeting via zinc finger proteins and CRISPR-Cas9. Notably, cRNAs bearing the encephalomyocarditis internal ribosome entry enabled robust expression and persistence compared with linear capped RNAs in cardiomyocytes and neurons, which highlights the utility of cRNAs in these non-dividing cells. We also describe genome targeting via deimmunized Cas9 delivered as cRNA and a long-range multiplexed protein engineering methodology for the combinatorial screening of deimmunized protein variants that enables compatibility between persistence of expression and immunogenicity in cRNA-delivered proteins. The cRNA toolset will aid research and the development of therapeutics.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests: The authors have filed patents based on this work. P.M. is a scientific co-founder of Shape Therapeutics, Navega Therapeutics, Pi Bio, Boundless Biosciences and Engine Biosciences. The terms of these arrangements have been reviewed and approved by the University of California, San Diego, in accordance with its conflict-of-interest policies. The other authors declare no competing interests.
Figures




References
-
- Kuhn AN et al. Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther. 17, 961–971 (2010). - PubMed
-
- Holtkamp S et al. Modification of antigen-encoding RNA increases stability, translational efficacy, and T-cell stimulatory capacity of dendritic cells. Blood 108, 4009–4017 (2006). - PubMed
MeSH terms
Substances
Grants and funding
- W81XWH-22-1-0401/U.S. Department of Defense (United States Department of Defense)
- R01 NS131560/NS/NINDS NIH HHS/United States
- AHA 916973/American Heart Association (American Heart Association, Inc.)
- P41 GM103504/GM/NIGMS NIH HHS/United States
- U54CA274502/U.S. Department of Health & Human Services | NIH | Office of Extramural Research, National Institutes of Health (OER)
- S10 OD026929/OD/NIH HHS/United States
- OT2 OD032742/OD/NIH HHS/United States
- DP2 NS111507/NS/NINDS NIH HHS/United States
- R01 HG012351/HG/NHGRI NIH HHS/United States
- U54 CA274502/CA/NCI NIH HHS/United States
- DP2NS111507/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- OT2OD032742/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
LinkOut - more resources
Full Text Sources
Research Materials

