Proteomic changes in Alzheimer's disease associated with progressive Aβ plaque and tau tangle pathologies
- PMID: 39187705
- PMCID: PMC11452344
- DOI: 10.1038/s41593-024-01737-w
Proteomic changes in Alzheimer's disease associated with progressive Aβ plaque and tau tangle pathologies
Abstract
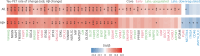
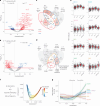
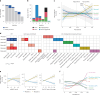
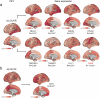
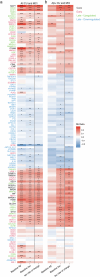
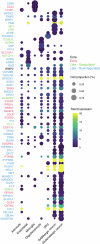
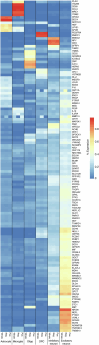
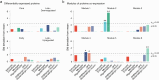
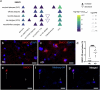
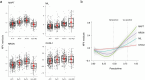
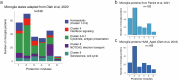
Proteomics can shed light on the dynamic and multifaceted alterations in neurodegenerative disorders like Alzheimer's disease (AD). Combining radioligands measuring β-amyloid (Aβ) plaques and tau tangles with cerebrospinal fluid proteomics, we uncover molecular events mirroring different stages of AD pathology in living humans. We found 127 differentially abundant proteins (DAPs) across the AD spectrum. The strongest Aβ-related proteins were mainly expressed in glial cells and included SMOC1 and ITGAM. A dozen proteins linked to ATP metabolism and preferentially expressed in neurons were independently associated with tau tangle load and tau accumulation. Only 20% of the DAPs were also altered in other neurodegenerative diseases, underscoring AD's distinct proteome. Two co-expression modules related, respectively, to protein metabolism and microglial immune response encompassed most DAPs, with opposing, staggered trajectories along the AD continuum. We unveil protein signatures associated with Aβ and tau proteinopathy in vivo, offering insights into complex neural responses and potential biomarkers and therapeutics targeting different disease stages.
© 2024. The Author(s).
Conflict of interest statement
O.H. has acquired research support (for the institution) from ADx, AVID Radiopharmaceuticals, Biogen, Eli Lilly, Eisai, Fujirebio, GE Healthcare, Pfizer and Roche. In the past 2 years, he has received consultancy/speaker fees from AC Immune, Amylyx, Alzpath, BioArctic, Biogen, Cerveau, Eisai, Eli Lilly, Fujirebio, Genentech, Merck, Novartis, Novo Nordisk, Roche, Sanofi and Siemens. S.P. has acquired research support (for the institution) from ki elements/ADDF. In the past 2 years, he has received consultancy/speaker fees from Bioartic, Biogen, Lilly and Roche. The remaining authors declare no competing interests.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

