Mechanism of structure-specific DNA binding by the FANCM branchpoint translocase
- PMID: 39189453
- PMCID: PMC11472164
- DOI: 10.1093/nar/gkae727
Mechanism of structure-specific DNA binding by the FANCM branchpoint translocase
Abstract
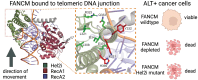
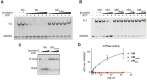
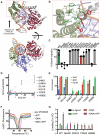
FANCM is a DNA repair protein that recognizes stalled replication forks, and recruits downstream repair factors. FANCM activity is also essential for the survival of cancer cells that utilize the Alternative Lengthening of Telomeres (ALT) mechanism. FANCM efficiently recognizes stalled replication forks in the genome or at telomeres through its strong affinity for branched DNA structures. In this study, we demonstrate that the N-terminal translocase domain drives this specific branched DNA recognition. The Hel2i subdomain within the translocase is crucial for effective substrate engagement and couples DNA binding to catalytic ATP-dependent branch migration. Removal of Hel2i or mutation of key DNA-binding residues within this domain diminished FANCM's affinity for junction DNA and abolished branch migration activity. Importantly, these mutant FANCM variants failed to rescue the cell cycle arrest, telomere-associated replication stress, or lethality of ALT-positive cancer cells depleted of endogenous FANCM. Our results reveal the Hel2i domain is key for FANCM to properly engage DNA substrates, and therefore plays an essential role in its tumour-suppressive functions by restraining the hyperactivation of the ALT pathway.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures





References
-
- Catucci I., Osorio A., Arver B., Neidhardt G., Bogliolo M., Zanardi F., Riboni M., Minardi S., Pujol R., Azzollini J.et al. .. Individuals with FANCM biallelic mutations do not develop Fanconi anemia, but show risk for breast cancer, chemotherapy toxicity and may display chromosome fragility. Genet. Med. 2018; 20:452–457. - PubMed
-
- Bogliolo M., Bluteau D., Lespinasse J., Pujol R., Vasquez N., Enghien C.D., Stoppa-Lyonnet D., Leblanc T., Soulier J., Surralles J.. Biallelic truncating FANCM mutations cause early-onset cancer but not Fanconi anemia. Genet. Med. 2018; 20:458–463. - PubMed
-
- Kiiski J.I., Pelttari L.M., Khan S., Freysteinsdottir E.S., Reynisdottir I., Hart S.N., Shimelis H., Vilske S., Kallioniemi A., Schleutker J.et al. .. Exome sequencing identifies FANCM as a susceptibility gene for triple-negative breast cancer. Proc. Natl. Acad. Sci. U.S.A. 2014; 111:15172–15177. - PMC - PubMed
-
- Basbous J., Constantinou A.. A tumor suppressive DNA translocase named FANCM. Crit. Rev. Biochem. Mol. Biol. 2019; 54:27–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

