SGLT2 inhibitor downregulated oxidative stress via activating AMPK pathway for cardiorenal (CR) protection in CR syndrome rodent fed with high protein diet
- PMID: 39190032
- PMCID: PMC11464616
- DOI: 10.1007/s10735-024-10233-1
SGLT2 inhibitor downregulated oxidative stress via activating AMPK pathway for cardiorenal (CR) protection in CR syndrome rodent fed with high protein diet
Abstract
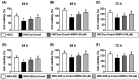
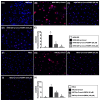
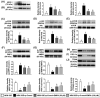
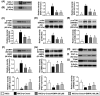
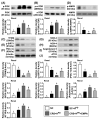
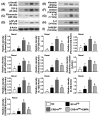
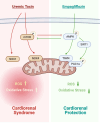
This study tested the hypothesis that empagliflozin (EMPA) therapy effectively protected renal and heart functions via downregulating reactive oxygen species (ROS) and activating AMPK signaling in cardiorenal syndrome (CRS) (induced by doxorubicin-5/6 nephrectomy) rats. In vitro result showed that underwent p-Cresol treatment, the H9C2/NRK-52E cell viabilities, were significantly suppressed, whereas cellular levels of ROS and early/late apoptosis of these cells were significantly increased that were significantly reversed by EMPA treatment (all p < 0.001). The protein levels of the cell-stress/oxidative signaling (p-PI3K/p-Akt/p-mTOR/NOXs/p-DRP1) were significantly activated, whereas the mitochondrial biogenesis signaling (p-AMPK/SIRT-1/TFAM/PGC-1α) was significantly repressed in these two cell lines treated by p-Cresol and all of these were significantly reversed by EMPA treatment (all p < 0.001). Male-adult-SD rats were categorized into groups 1 [sham-operated control (SC)]/2 [SC + high protein diet (HPD) since day 1 after CKD induction]/3 (CRS + HPD)/4 (CRS + HPD+EMPA/20 mg/kg/day) and heart/kidney were harvested by day 60. By day 63, the renal function parameters (creatinine/BUN/proteinuria)/renal artery restrictive index/cellular levels of ROS/inflammation were significantly increased in group 3 than in groups 1/2, whereas heart function exhibited an opposite pattern of ROS among the groups, and all of these parameters were significantly reversed by EMPA treatment (all p < 0.0001). The protein levels of inflammation/ oxidative-stress/cell-stress signalings were highest in group 2, lowest in group 1 and significantly lower in group 4 than in group 2, whereas the AMPK-mitochondrial biogenesis displayed an opposite manner of oxidative-stress among the groups (all p < 0.0001). EMPA treatment effectively protected the heart/kidney against CRS damage via suppressing ROS signaling and upregulating AMPK-mediated mitochondrial biogenesis.
Keywords: Cardiorenal syndrome; Empagliflozin; Inflammation; Mitochondrial biogenesis; Oxidative stress.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

