Synonymous codon substitutions modulate transcription and translation of a divergent upstream gene by modulating antisense RNA production
- PMID: 39190361
- PMCID: PMC11388325
- DOI: 10.1073/pnas.2405510121
Synonymous codon substitutions modulate transcription and translation of a divergent upstream gene by modulating antisense RNA production
Abstract
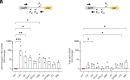
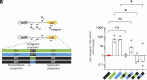
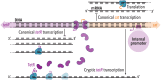
Synonymous codons were originally viewed as interchangeable, with no phenotypic consequences. However, substantial evidence has now demonstrated that synonymous substitutions can perturb a variety of gene expression and protein homeostasis mechanisms, including translational efficiency, translational fidelity, and cotranslational folding of the encoded protein. To date, most studies of synonymous codon-derived perturbations have focused on effects within a single gene. Here, we show that synonymous codon substitutions made far within the coding sequence of Escherichia coli plasmid-encoded chloramphenicol acetyltransferase (cat) can significantly increase expression of the divergent upstream tetracycline resistance gene, tetR. In four out of nine synonymously recoded cat sequences tested, expression of the upstream tetR gene was significantly elevated due to transcription of a long antisense RNA (asRNA) originating from a transcription start site within cat. Surprisingly, transcription of this asRNA readily bypassed the native tet transcriptional repression mechanism. Even more surprisingly, accumulation of the TetR protein correlated with the level of asRNA, rather than total tetR RNA. These effects of synonymous codon substitutions on transcription and translation of a neighboring gene suggest that synonymous codon usage in bacteria may be under selection to both preserve the amino acid sequence of the encoded gene and avoid DNA sequence elements that can significantly perturb expression of neighboring genes. Avoiding such sequences may be especially important in plasmids and prokaryotic genomes, where genes and regulatory elements are often densely packed. Similar considerations may apply to the design of genetic circuits for synthetic biology applications.
Keywords: antisense RNA; gene expression; overlapping divergent promoters; protein homeostasis; synonymous codon substitutions.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures





Comment in
-
Secret code: Encoding promoters by synonymous codons.Proc Natl Acad Sci U S A. 2024 Oct;121(40):e2416360121. doi: 10.1073/pnas.2416360121. Epub 2024 Sep 23. Proc Natl Acad Sci U S A. 2024. PMID: 39312671 Free PMC article. No abstract available.
References
-
- Chaney J. L., Clark P. L., Roles for synonymous codon usage in protein biogenesis. Ann. Rev. Biophys. 44, 143–166 (2015). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

