Community assessment of methods to deconvolve cellular composition from bulk gene expression
- PMID: 39191725
- PMCID: PMC11350143
- DOI: 10.1038/s41467-024-50618-0
Community assessment of methods to deconvolve cellular composition from bulk gene expression
Erratum in
-
Author Correction: Community assessment of methods to deconvolve cellular composition from bulk gene expression.Nat Commun. 2024 Nov 12;15(1):9783. doi: 10.1038/s41467-024-53843-9. Nat Commun. 2024. PMID: 39532851 Free PMC article. No abstract available.
Abstract
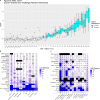
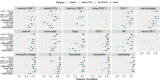
We evaluate deconvolution methods, which infer levels of immune infiltration from bulk expression of tumor samples, through a community-wide DREAM Challenge. We assess six published and 22 community-contributed methods using in vitro and in silico transcriptional profiles of admixed cancer and healthy immune cells. Several published methods predict most cell types well, though they either were not trained to evaluate all functional CD8+ T cell states or do so with low accuracy. Several community-contributed methods address this gap, including a deep learning-based approach, whose strong performance establishes the applicability of this paradigm to deconvolution. Despite being developed largely using immune cells from healthy tissues, deconvolution methods predict levels of tumor-derived immune cells well. Our admixed and purified transcriptional profiles will be a valuable resource for developing deconvolution methods, including in response to common challenges we observe across methods, such as sensitive identification of functional CD4+ T cell states.
© 2024. The Author(s).
Conflict of interest statement
A.M.N. is a co-founder of CiberMed, Inc., and A.J.G. has consulted for CiberMed, Inc. J.S.R. received funding from GlaxoSmithKline and Sanofi and consultant fees from Travere Therapeutics and Astex Therapeutic. A.V. is currently employed by F. Hoffmann-La Roche Ltd. The remaining authors declare no competing interests.
Figures





References
MeSH terms
Grants and funding
- U24 CA209923/CA/NCI NIH HHS/United States
- R01 GM122085/GM/NIGMS NIH HHS/United States
- CA209971/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R01GM122085/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U24CA224309/U.S. Department of Health & Human Services | National Institutes of Health (NIH)
- U54 CA209971/CA/NCI NIH HHS/United States
- CA209923/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- U24 CA224309/CA/NCI NIH HHS/United States
- U54 CA209988/CA/NCI NIH HHS/United States
- CA209988/U.S. Department of Health & Human Services | NIH | National Cancer Institute (NCI)
- R35 GM149261/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

