SREBP2 restricts osteoclast differentiation and activity by regulating IRF7 and limits inflammatory bone erosion
- PMID: 39191742
- PMCID: PMC11350122
- DOI: 10.1038/s41413-024-00354-4
SREBP2 restricts osteoclast differentiation and activity by regulating IRF7 and limits inflammatory bone erosion
Abstract
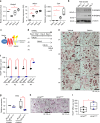
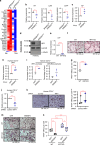
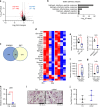
Osteoclasts are multinucleated bone-resorbing cells, and their formation is tightly regulated to prevent excessive bone loss. However, the mechanisms by which osteoclast formation is restricted remain incompletely determined. Here, we found that sterol regulatory element binding protein 2 (SREBP2) functions as a negative regulator of osteoclast formation and inflammatory bone loss. Cholesterols and SREBP2, a key transcription factor for cholesterol biosynthesis, increased in the late phase of osteoclastogenesis. The ablation of SREBP2 in myeloid cells resulted in increased in vivo and in vitro osteoclastogenesis, leading to low bone mass. Moreover, deletion of SREBP2 accelerated inflammatory bone destruction in murine inflammatory osteolysis and arthritis models. SREBP2-mediated regulation of osteoclastogenesis is independent of its canonical function in cholesterol biosynthesis but is mediated, in part, by its downstream target, interferon regulatory factor 7 (IRF7). Taken together, our study highlights a previously undescribed role of the SREBP2-IRF7 regulatory circuit as a negative feedback loop in osteoclast differentiation and represents a novel mechanism to restrain pathological bone destruction.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
- R01 AR073156/AR/NIAMS NIH HHS/United States
- AR073156/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- AR069562/U.S. Department of Health & Human Services | NIH | National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS)
- P30 CA008748/CA/NCI NIH HHS/United States
- 2020R1A6A3A03037133/National Research Foundation of Korea (NRF)
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

