Antibody Fc receptor binding and T cell responses to homologous and heterologous immunization with inactivated or mRNA vaccines against SARS-CoV-2
- PMID: 39191745
- PMCID: PMC11350167
- DOI: 10.1038/s41467-024-51427-1
Antibody Fc receptor binding and T cell responses to homologous and heterologous immunization with inactivated or mRNA vaccines against SARS-CoV-2
Abstract
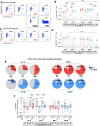
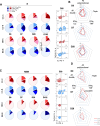
Whole virion inactivated vaccine CoronaVac (C) and Spike (S) mRNA BNT162b2 (B) vaccines differ greatly in their ability to elicit neutralizing antibodies but have somewhat comparable effectiveness in protecting from severe COVID-19. We conducted further analyses for a randomized trial (Cobovax study, NCT05057169) of third dose homologous and heterologous booster vaccination, i.e. four interventions CC-C, CC-B, BB-C and BB-B. Here, we assess vaccine immunogenicity beyond neutralizing function, including S and non-S antibodies with Fc receptor (FcR) binding, antibody avidity and T cell specificity to 6 months post-vaccination. Ancestral and Omicron S-specific IgG and FcR binding are significantly higher by BNT162b2 booster than CoronaVac, regardless of first doses. Nucleocapsid (N) antibodies are only increased in homologous boosted CoronaVac participants (CC-C). CoronaVac primed participants have lower baseline S-specific CD4+ IFNγ+ cells, but are significantly increased by either CoronaVac or BNT162b2 boosters. Priming vaccine content defined T cell peptide specificity preference, with S-specific T cells dominating B primed groups and non-S structural peptides contributing more in C primed groups, regardless of booster type. S-specific CD4+ T cell responses, N-specific antibodies, and antibody effector functions via Fc receptor binding may contribute to protection and compensate for less potent neutralizing responses in CoronaVac recipients.
© 2024. The Author(s).
Conflict of interest statement
BJC has consulted for AstraZeneca, Fosun Pharma, GSK, Haleon, Moderna, Roche, and Sanofi Pasteur. LMP, JSMP, SAV have patented (2021800455096 and 17/997,434) for the use of ORF8 as diagnostics of SARS-CoV-2 infection. MM produced ORF8 by patent process based on US Patents 8,507,220 and 8,586,826. The authors report no other potential conflicts of interest.
Figures





References
-
- Tsang, N. N. Y., So, H. C., Cowling, B. J., Leung, G. M. & Ip, D. K. M. Effectiveness of BNT162b2 and CoronaVac COVID-19 vaccination against asymptomatic and symptomatic infection of SARS-CoV-2 omicron BA.2 in Hong Kong: a prospective cohort study. Lancet Infect Dis23, 421–434 (2023). 10.1016/S1473-3099(22)00732-0 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

