Galectins induced from hemocytes bridge phosphatidylserine and N-glycosylated Drpr/CED-1 receptor during dendrite pruning
- PMID: 39191750
- PMCID: PMC11349873
- DOI: 10.1038/s41467-024-51581-6
Galectins induced from hemocytes bridge phosphatidylserine and N-glycosylated Drpr/CED-1 receptor during dendrite pruning
Abstract
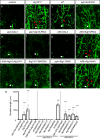
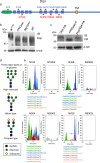
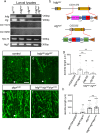
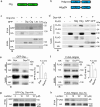
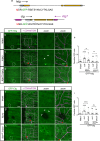
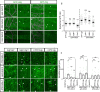
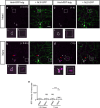
During neuronal pruning, phagocytes engulf shed cellular debris to avoid inflammation and maintain tissue homeostasis. How phagocytic receptors recognize degenerating neurites had been unclear. Here, we identify two glucosyltransferases Alg8 and Alg10 of the N-glycosylation pathway required for dendrite fragmentation and clearance through genetic screen. The scavenger receptor Draper (Drpr) is N-glycosylated with complex- or hybrid-type N-glycans that interact specifically with galectins. We also identify the galectins Crouching tiger (Ctg) and Hidden dragon (Hdg) that interact with N-glycosylated Drpr and function in dendrite pruning via the Drpr pathway. Ctg and Hdg are required in hemocytes for expression and function, and are induced during dendrite injury to localize to injured dendrites through specific interaction with exposed phosphatidylserine (PS) on the surface membrane of injured dendrites. Thus, the galectins Ctg and Hdg bridge the interaction between PS and N-glycosylated Drpr, leading to the activation of phagocytosis.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
The Drosophila chemokine-like Orion bridges phosphatidylserine and Draper in phagocytosis of neurons.Proc Natl Acad Sci U S A. 2023 Jun 13;120(24):e2303392120. doi: 10.1073/pnas.2303392120. Epub 2023 Jun 5. Proc Natl Acad Sci U S A. 2023. PMID: 37276397 Free PMC article.
-
Epidermal cells are the primary phagocytes in the fragmentation and clearance of degenerating dendrites in Drosophila.Neuron. 2014 Feb 5;81(3):544-560. doi: 10.1016/j.neuron.2013.11.021. Epub 2014 Jan 9. Neuron. 2014. PMID: 24412417 Free PMC article.
-
Draper-mediated and phosphatidylserine-independent phagocytosis of apoptotic cells by Drosophila hemocytes/macrophages.J Biol Chem. 2004 Nov 12;279(46):48466-76. doi: 10.1074/jbc.M408597200. Epub 2004 Sep 1. J Biol Chem. 2004. PMID: 15342648
-
Glycosylation, galectins and cellular signaling.Curr Opin Cell Biol. 2011 Aug;23(4):383-92. doi: 10.1016/j.ceb.2011.05.001. Epub 2011 May 26. Curr Opin Cell Biol. 2011. PMID: 21616652 Review.
-
Spatiotemporal regulation of developmental neurite pruning: Molecular and cellular insights from Drosophila models.Neurosci Res. 2021 Jun;167:54-63. doi: 10.1016/j.neures.2020.11.010. Epub 2020 Dec 10. Neurosci Res. 2021. PMID: 33309868 Review.
Cited by
-
Biochemical and biophysical mechanisms macrophages use to tune phagocytic appetite.J Cell Sci. 2025 Jan 1;138(1):JCS263513. doi: 10.1242/jcs.263513. Epub 2025 Jan 3. J Cell Sci. 2025. PMID: 39749603 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

