RNA nanotherapeutics with fibrosis overexpression and retention for MASH treatment
- PMID: 39191801
- PMCID: PMC11350072
- DOI: 10.1038/s41467-024-51571-8
RNA nanotherapeutics with fibrosis overexpression and retention for MASH treatment
Abstract
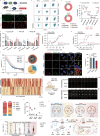
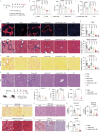
Metabolic dysfunction-associated steatohepatitis (MASH) poses challenges for targeted delivery and retention of therapeutic proteins due to excess extracellular matrix (ECM). Here we present a new approach to treat MASH, termed "Fibrosis overexpression and retention (FORT)". In this strategy, we design (1) retinoid-derivative lipid nanoparticle (LNP) to enable enhanced mRNA overexpression in fibrotic regions, and (2) mRNA modifications which facilitate anchoring of therapeutic proteins in ECM. LNPs containing carboxyl-retinoids, rather than alcohol- or ester-retinoids, effectively deliver mRNA with over 10-fold enhancement of protein expression in fibrotic livers. The carboxyl-retinoid rearrangement on the LNP surface improves protein binding and membrane fusion. Therapeutic proteins are then engineered with an endogenous collagen-binding domain. These fusion proteins exhibit increased retention in fibrotic lesions and reduced systemic toxicity. In vivo, fibrosis-targeting LNPs encoding fusion proteins demonstrate superior therapeutic efficacy in three clinically relevant male-animal MASH models. This approach holds promise in fibrotic diseases unsuited for protein injection.
© 2024. The Author(s).
Conflict of interest statement
M, X.Z.S., Z.Q.Z. have filed a patent for the development of the described FORT strategy. The remaining authors declare no competing interests.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

