A cell state-specific metabolic vulnerability to GPX4-dependent ferroptosis in glioblastoma
- PMID: 39192032
- PMCID: PMC11480389
- DOI: 10.1038/s44318-024-00176-4
A cell state-specific metabolic vulnerability to GPX4-dependent ferroptosis in glioblastoma
Abstract
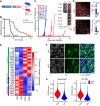
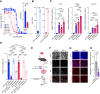
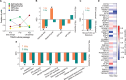
Glioma cells hijack developmental programs to control cell state. Here, we uncover a glioma cell state-specific metabolic liability that can be therapeutically targeted. To model cell conditions at brain tumor inception, we generated genetically engineered murine gliomas, with deletion of p53 alone (p53) or with constitutively active Notch signaling (N1IC), a pathway critical in controlling astrocyte differentiation during brain development. N1IC tumors harbored quiescent astrocyte-like transformed cell populations while p53 tumors were predominantly comprised of proliferating progenitor-like cell states. Further, N1IC transformed cells exhibited increased mitochondrial lipid peroxidation, high ROS production and depletion of reduced glutathione. This altered mitochondrial phenotype rendered the astrocyte-like, quiescent populations more sensitive to pharmacologic or genetic inhibition of the lipid hydroperoxidase GPX4 and induction of ferroptosis. Treatment of patient-derived early-passage cell lines and glioma slice cultures generated from surgical samples with a GPX4 inhibitor induced selective depletion of quiescent astrocyte-like glioma cell populations with similar metabolic profiles. Collectively, these findings reveal a specific therapeutic vulnerability to ferroptosis linked to mitochondrial redox imbalance in a subpopulation of quiescent astrocyte-like glioma cells resistant to standard forms of treatment.
Keywords: Astrocytic; Ferroptosis; Glioma; Mitochondrial-metabolism; Quiescent.
© 2024. The Author(s).
Figures




Update of
-
A cell state specific metabolic vulnerability to GPX4-dependent ferroptosis in glioblastoma.bioRxiv [Preprint]. 2023 Feb 23:2023.02.22.529581. doi: 10.1101/2023.02.22.529581. bioRxiv. 2023. Update in: EMBO J. 2024 Oct;43(20):4492-4521. doi: 10.1038/s44318-024-00176-4. PMID: 36865302 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- R38 CA231577/CA/NCI NIH HHS/United States
- R35 CA209896/CA/NCI NIH HHS/United States
- P30CA013696/HHS | NIH | National Cancer Institute (NCI)
- T32 GM145440/GM/NIGMS NIH HHS/United States
- P30 CA013696/CA/NCI NIH HHS/United States
- R01 HL112626/HL/NHLBI NIH HHS/United States
- R01HL112626/HHS | National Institutes of Health (NIH)
- R01 NS103473/NS/NINDS NIH HHS/United States
- U01 CA272610/CA/NCI NIH HHS/United States
- R01NS103473/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R35CA209896/HHS | National Institutes of Health (NIH)
- R01 HL062454/HL/NHLBI NIH HHS/United States
- S10 OD032433/OD/NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

