STIC2 selectively binds ribosome-nascent chain complexes in the cotranslational sorting of Arabidopsis thylakoid proteins
- PMID: 39192033
- PMCID: PMC11480477
- DOI: 10.1038/s44318-024-00211-4
STIC2 selectively binds ribosome-nascent chain complexes in the cotranslational sorting of Arabidopsis thylakoid proteins
Abstract
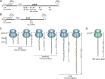
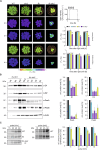
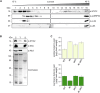
Chloroplast-encoded multi-span thylakoid membrane proteins are crucial for photosynthetic complexes, yet the coordination of their biogenesis remains poorly understood. To identify factors that specifically support the cotranslational biogenesis of the reaction center protein D1 of photosystem (PS) II, we generated and affinity-purified stalled ribosome-nascent chain complexes (RNCs) bearing D1 nascent chains. Stalled RNCs translating the soluble ribosomal subunit uS2c were used for comparison. Quantitative tandem-mass spectrometry of the purified RNCs identified around 140 proteins specifically associated with D1 RNCs, mainly involved in protein and cofactor biogenesis, including chlorophyll biosynthesis, and other metabolic pathways. Functional analysis of STIC2, a newly identified D1 RNC interactor, revealed its cooperation with chloroplast protein SRP54 in the de novo biogenesis and repair of D1, and potentially other cotranslationally-targeted reaction center subunits of PSII and PSI. The primary binding interface between STIC2 and the thylakoid insertase Alb3 and its homolog Alb4 was mapped to STIC2's β-sheet region, and the conserved Motif III in the C-terminal regions of Alb3/4.
Keywords: Arabidopsis; Cotranslational Targeting; Photosystem II; STIC2; Thylakoid Membrane.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

